webSDA example: simulation of diffusion of multiple molecules for lysozyme
The "SDA multiple molecules" module is used to simulate the diffusional motion of several hundred macromolecules in a periodic box. In this example, "SDA multiple molecules" (SDAMM) is applied to lysozyme (at pH 6).
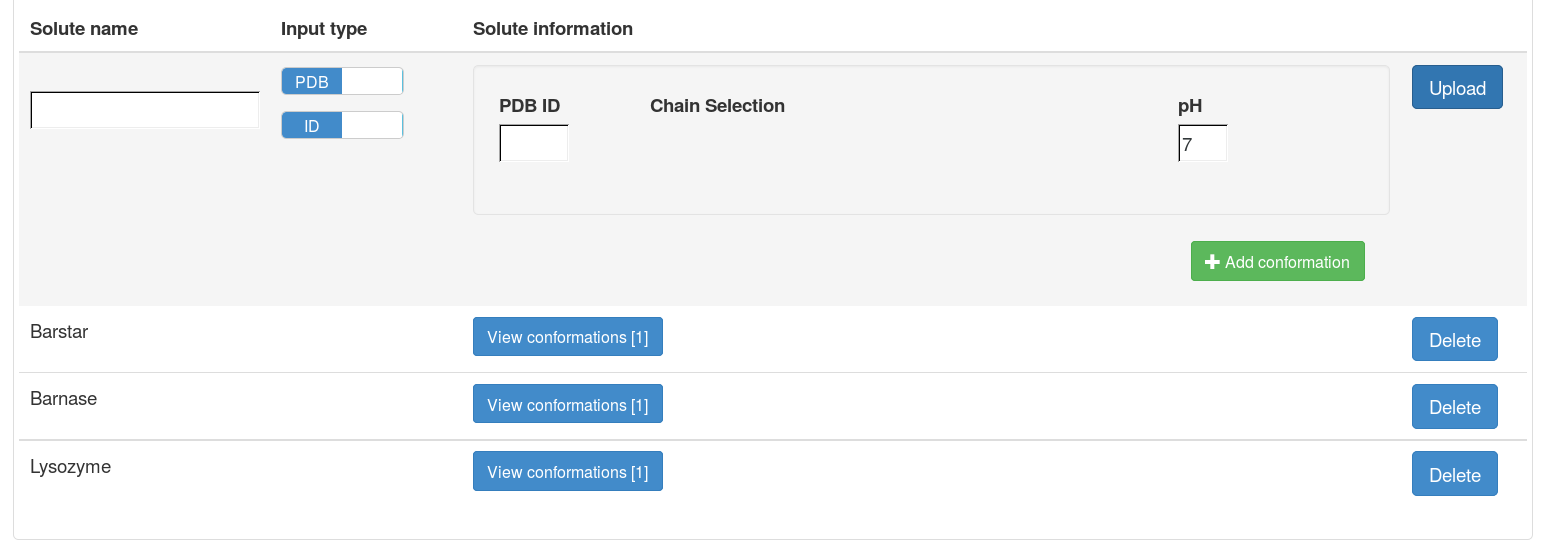
After uploading each file, the information on the uploaded solute will be shown on the right side of the file upload panel, including the name of the solute and the number of PQR files for this solute (number of conformations). A solute can only be deleted when it is not associated with any projects.
Now, choose the "SDA multiple molecules" method:
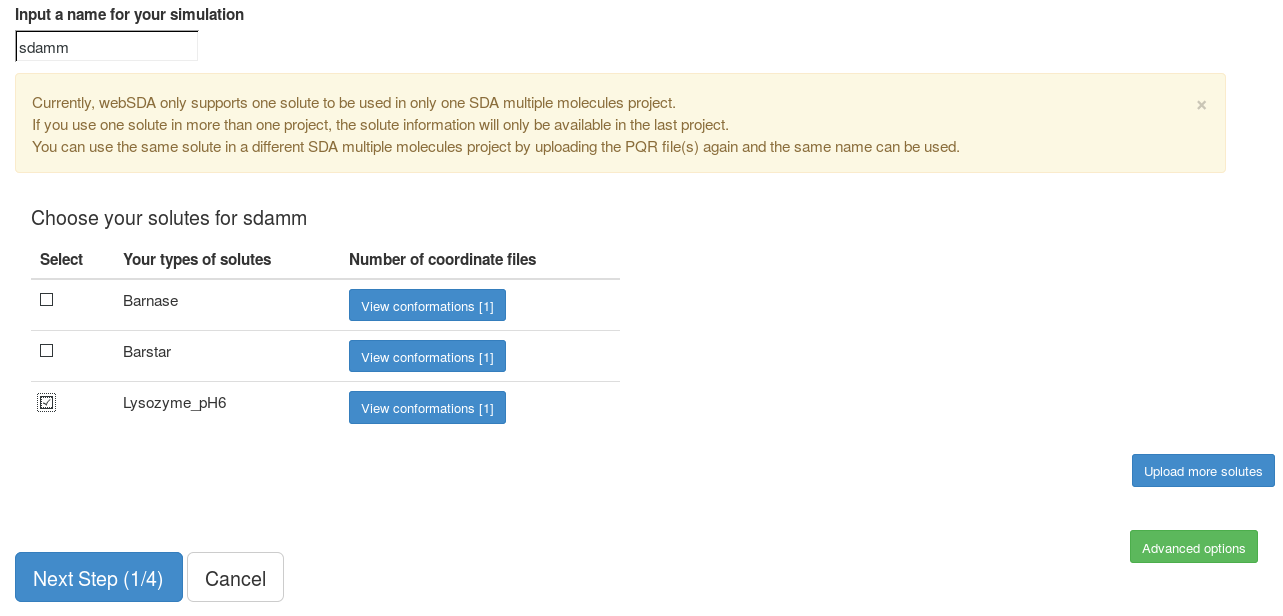
This goes to the page to set up a new SDA multiple molecules project. Note: currently, one solute can only be used in one SDA multiple molecules project. If the same solute is used in a new project, this solute will be deleted from the previous project. To use the same solute in a new project, it is better to upload this solute again. The same name can be used for this solute.
Here, we choose "Lysozyme_ph6" as our solute. We use default parameters in the "Advanced options".
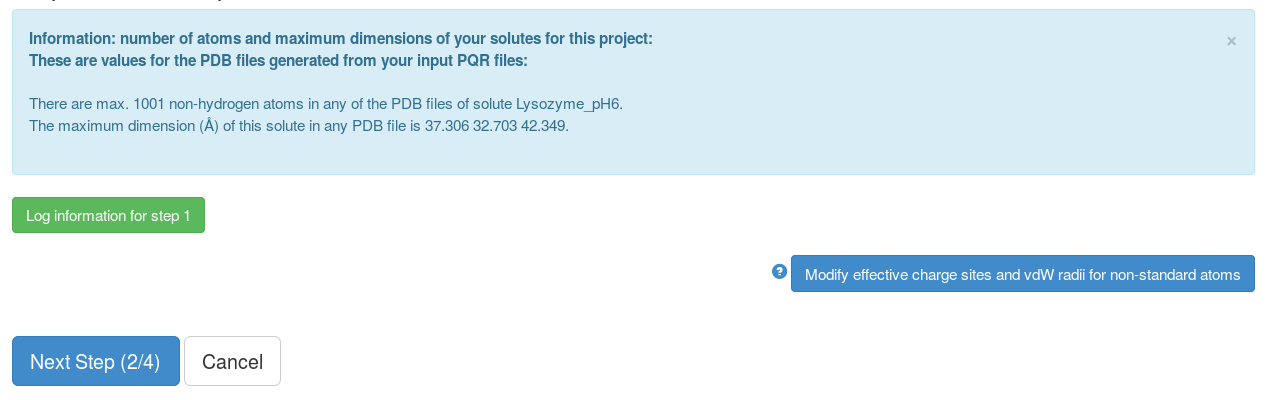
A python script in the background checks the PQR files, generates necessary information (net charge, center of geometry, molecular weight etc.) and input files (to calculate the interaction terms).
This page shows some of the information from the previous step, non-standard atoms whose effective charges sites (normally on ligands) are not known. We click "Next Step (2/4)" and after that, the grid files for the interaction terms (here electrostatic interaction and non-polar forces) for each solute will be computed.
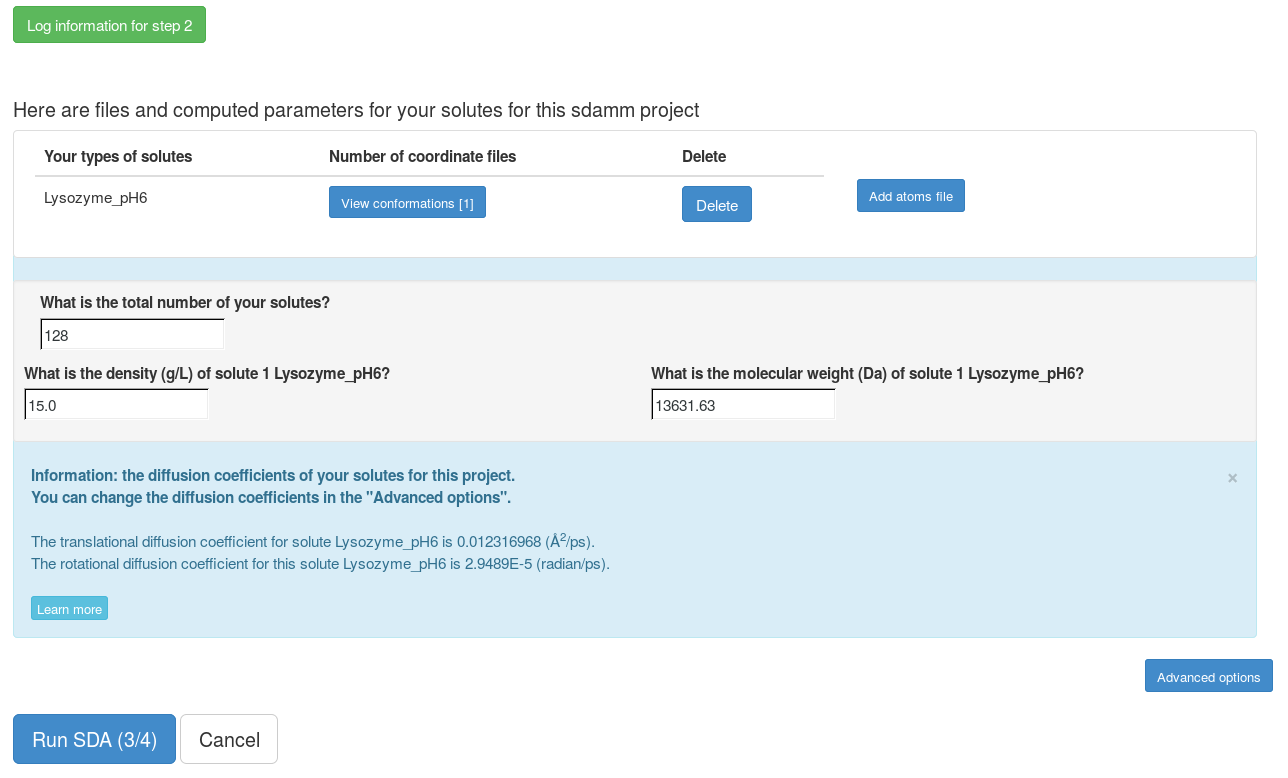
Now, the grid files for the interactions for each solute have been computed. We can modify the number of solutes that we will simulate and the density of the solutes in our simulation. In the "Advanced options", the SDA input file can be generated and all the files can be downloaded to run the SDA7 standalone. The web server can only be used to run short SDA jobs. To get more accurate results, these input files should be downloaded and used to run the standalone SDA7.
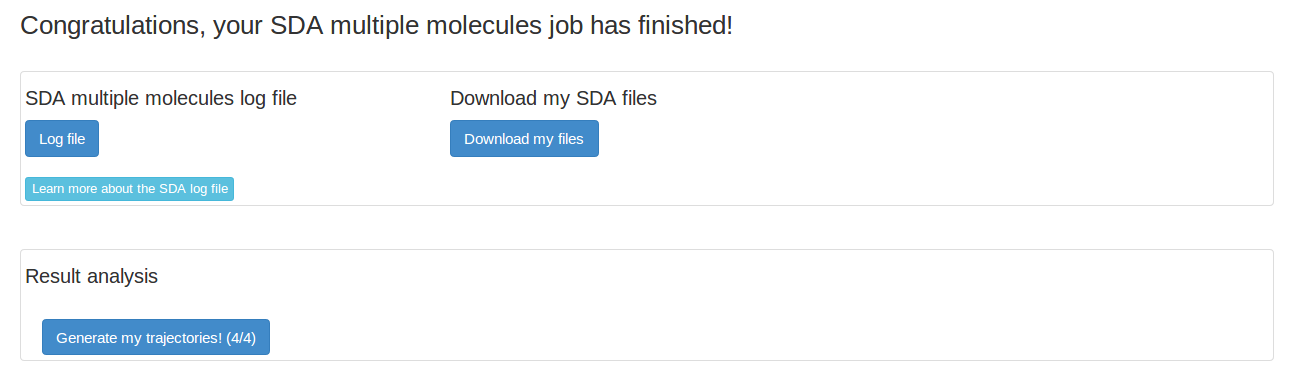
We submit the SDA job with default parameters. For this lysozyme SDAMM simulation, it takes about 2 minutes.
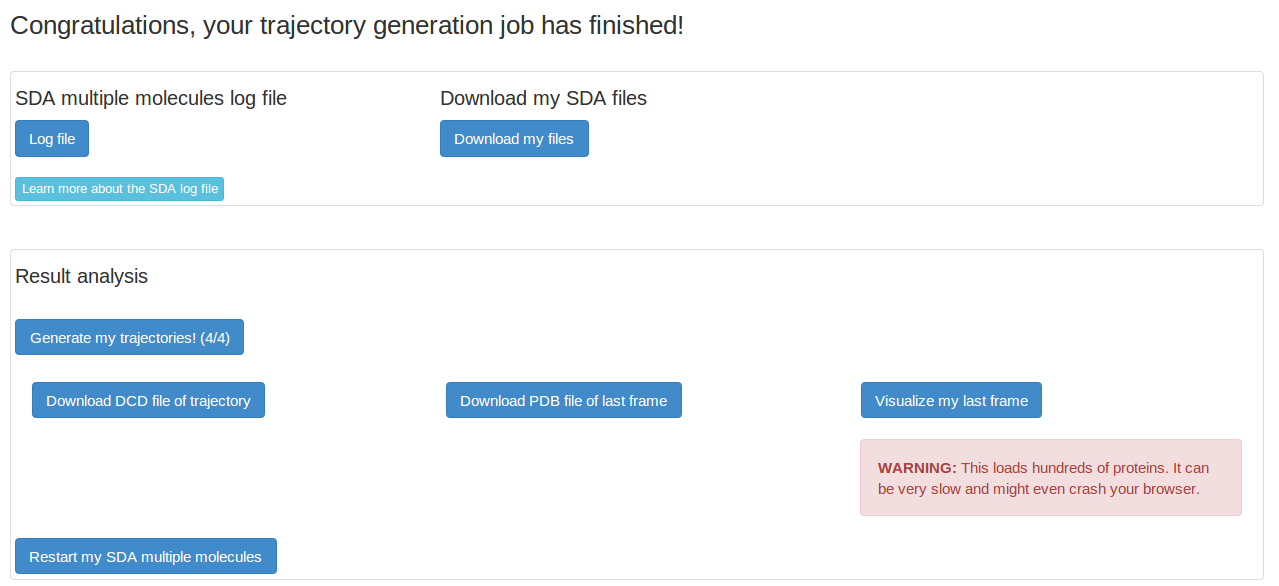
The SDA log file gives detailed information on the SDA run. We want to look at our trajectory, so we need to first generate the trajectory.
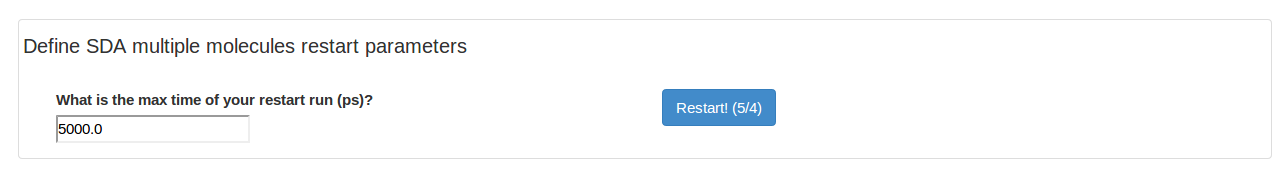
This page shows the SDA multiple molecules trajectory and the last frame of the trajectory in PDB format. This page also contains the option to restart the current SDA multiple molecules trajectory. Note, only one restart will be allowed in webSDA!
We can extend our trajectory to a longer time scale. It is recommended to download the SDA files and run the standalone SDA7 locally using e.g. VMD.
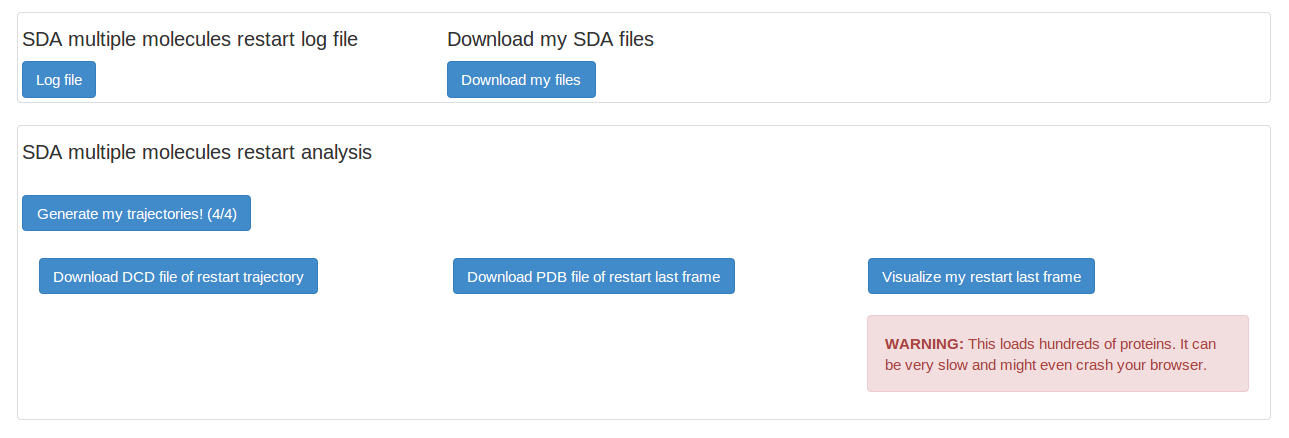
The restart of our multiple molecules project produces similar output files. We can generate SDA trajectory and PDB file of the last frame the same as we did for the first SDA run.
We can have a look at our last frame of the SDA multiple molecules trajectory. However, we are loading hundreds of proteins into our browser and this is normally very slow. It is always recommended to download the dcd trajectory file and the PDB file of the last frame and visualize them locally.