in silico tethering 4
(E. coli TS-mutant
L143C,C146S)
Tethering with dimer of E. coli
TS (1f4d.pdb, chain A and B, L143C, C146S)
- download 1f4d.pdb
- superimpose to 1hvy-AB_addH.wi.pdb
(sup2pdbs_sh
1hvy-AB_addH.wi.pdb 1f4d.pdb)
- 1f4d.pdbf
- load in pymol, correct bonds of ligand and addH - 1f4d_addH.wat.pdb
- protonation with whatif using water from X-ray structure (whatif.sh
1f4d_addh.wat) - 1f4d_addh.wat.wi.pdb
- remove water molecules but keeping sulfate - 1f4d_addh.wi.pdb
- load 1f4d_addh.wi.pdb in
pymol and save again
(this step is important otherwise GOLD has problems with
atom types)
- remove ligand and side chain of C143
in monomer A using pymol -1f4d_addh-L143C.sulf.wi.pdb
- center Ca-C143: 2300
(center 1hvy-Ca-C195: 3.002
2.973 11.352)
radius: 25 A
linking atom of receptor 1f4d_addh-L143C.sulf.wi.pdb:
2300
linking atom of ligand CID_445503-cys3D.sdf:
43
- start GOLD 3.1.1: gold_di1f4cL143C_wi.sulf.conf
- WARNING: problems with atom types in mol2
output: docking_di1f4dL143C-sulf_wi_445503.25A.sdf
- active_atoms.pdb
active_atoms.1f4d_addh-L143C.wi.pdb
- gold_protein.mol2
gold_protein.1f4d_addh-L143C.wi.mol2
/home/henricsn/LIGHTS/results/docking3_C195/
GoldScore
gold_analysis.sh -xray1
1f4d_CID_445503-cys3D.pdb -file
docking_di1f4dL143C-sulf_wi_445503.25A.sdf
# res RMSD Fit iVDW iTOR eVDW iHB eHB covENE ENE iVDWw eVDWw iHBw eHBw covENEw
1 xray 2.657 -68.1240 1.5248 -6.1141 -43.0613 0.0000 0.0000 -4.3253 1.5248 -6.1141 -59.2093 0.0000 0.0000 -4.3253
2 xray 4.033 -69.2604 1.6532 -7.6193 -45.1297 0.0000 1.3901 -2.6310 1.6532 -7.6193 -62.0533 0.0000 1.3901 -2.6310
3 xray 5.859 -69.8210 3.6317 -6.8875 -46.6300 0.0000 0.3918 -2.8408 3.6317 -6.8875 -64.1163 0.0000 0.3918 -2.8408
4 xray 3.469 -70.9946 4.0834 -5.0868 -49.5251 0.0000 2.0915 -3.9858 4.0834 -5.0868 -68.0970 0.0000 2.0915 -3.9858
5 xray 4.008 -71.0893 1.4745 -6.8307 -45.3323 0.0000 2.0000 -5.4012 1.4745 -6.8307 -62.3319 0.0000 2.0000 -5.4012
6 xray 7.072 -71.2313 -2.0349 -8.0191 -41.4681 0.0000 0.0000 -4.1587 -2.0349 -8.0191 -57.0186 0.0000 0.0000 -4.1587
7 xray 4.925 -71.4415 1.0719 -8.3121 -45.3012 0.0000 0.9897 -2.9018 1.0719 -8.3121 -62.2892 0.0000 0.9897 -2.9018
8 xray 5.267 -72.7463 2.1361 -7.1940 -47.7296 0.0000 0.9545 -3.0148 2.1361 -7.1940 -65.6281 0.0000 0.9545 -3.0148
9 xray 2.729 -74.0007 0.9326 -8.2060 -44.7993 0.0000 0.0000 -5.1283 0.9326 -8.2060 -61.5990 0.0000 0.0000 -5.1283
10 xray 1.948 -75.3501 0.3136 -5.3347 -48.6601 0.0000 0.1146 -3.5359 0.3136 -5.3347 -66.9077 0.0000 0.1146 -3.5359
ChemScore
gold_analysis.sh -xray1 1f4d_CID_445503-cys3D.pdb -file
docking_di1f4dL143C-sulf_wi_445503.CS25A.sdf
# res RMSD Fit DG ZeCo HB MET LIPO iHB DEC DEI ROT COVA HBw METw LIPOw iHBw DECw DEIw ROTw
1 xray 2.659 31.7229 -31.5104 -5.4800 1.9217 0.0000 202.3888 0.0000 -4.4855 3.1630 1.5889 1.1100 -6.4185 -0.0000 -23.6795 0.0000 -4.4855 3.1630 4.0676
2 xray 2.583 30.8349 -30.3043 -5.4800 1.8140 0.0000 195.1552 0.0000 -3.7850 2.9623 1.5889 0.2921 -6.0587 -0.0000 -22.8332 0.0000 -3.7850 2.9623 4.0676
3 xray 2.750 30.8128 -30.9784 -5.4800 1.9081 0.0000 198.2287 0.0000 -4.7075 3.7748 1.5889 1.0983 -6.3732 -0.0000 -23.1928 0.0000 -4.7075 3.7748 4.0676
4 xray 3.124 29.5037 -28.5815 -5.4800 1.7310 0.0000 182.8008 0.0000 -4.5363 2.9088 1.5889 0.7052 -5.7814 -0.0000 -21.3877 0.0000 -4.5363 2.9088 4.0676
5 xray 2.579 29.4834 -29.0477 -5.4800 1.5471 0.0000 192.0349 0.0000 -4.0053 2.8335 1.5889 0.7361 -5.1672 -0.0000 -22.4681 0.0000 -4.0053 2.8335 4.0676
6 xray 2.709 28.8331 -30.7201 -5.4800 1.6936 0.0000 202.1469 0.0000 -2.3418 3.8629 1.5889 0.3659 -5.6565 -0.0000 -23.6512 0.0000 -2.3418 3.8629 4.0676
7 xray 2.683 28.6245 -28.9974 -5.4800 0.9670 0.0000 208.1648 0.0000 -3.1637 3.2314 1.5889 0.3052 -3.2297 -0.0000 -24.3553 0.0000 -3.1637 3.2314 4.0676
8 xray 3.353 26.7041 -25.5758 -5.4800 0.9994 0.0000 177.9957 0.0000 -4.4579 3.1783 1.5889 0.1513 -3.3379 -0.0000 -20.8255 0.0000 -4.4579 3.1783 4.0676
9 xray 3.527 26.1224 -24.9551 -5.4800 1.0000 0.0000 172.6725 0.0000 -4.4602 3.2275 1.5889 0.0653 -3.3399 -0.0000 -20.2027 0.0000 -4.4602 3.2275 4.0676
10 xray 3.249 24.9502 -25.4633 -5.4800 0.9999 0.0000 177.0203 0.0000 -3.5225 3.3091 1.5889 0.7265 -3.3395 -0.0000 -20.7114 0.0000 -3.5225 3.3091 4.0676
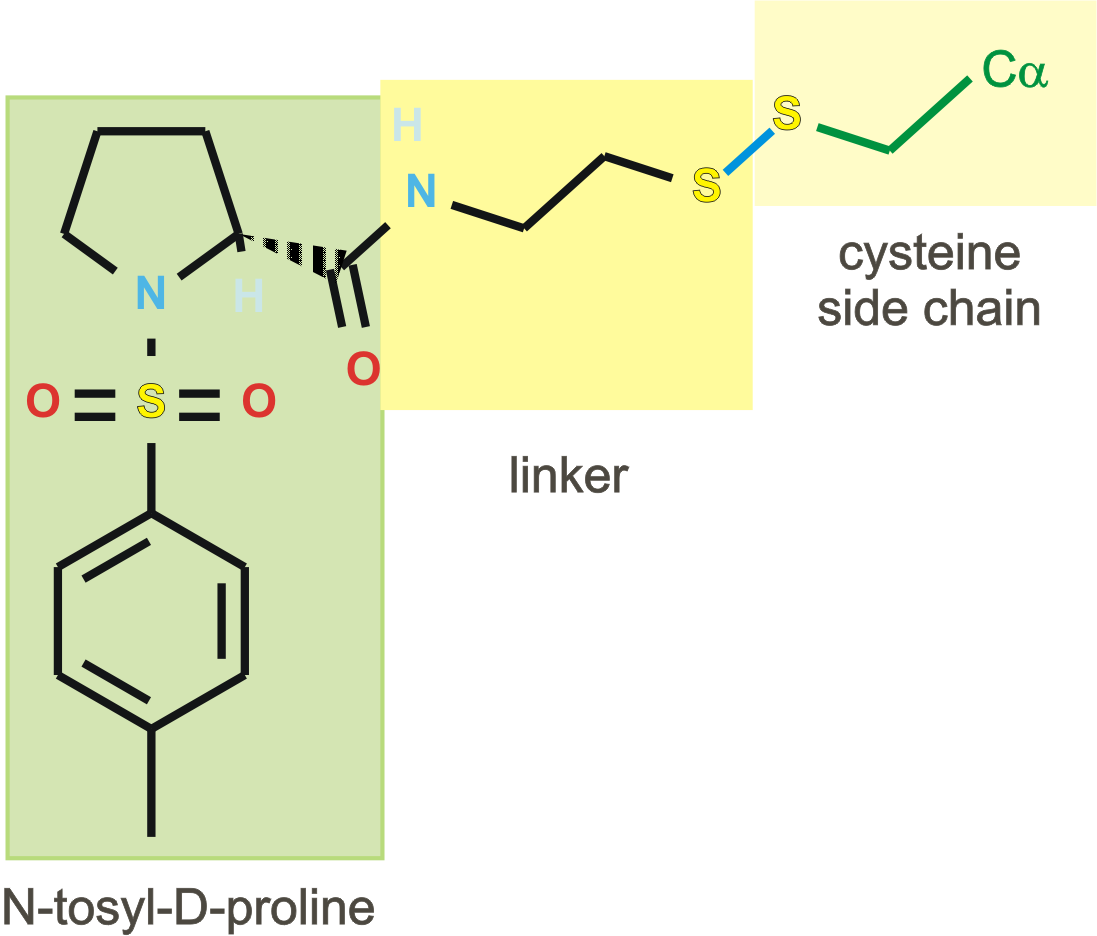
In contrast to 1f4c, 1f4f and 1f4g, 1f4d containts the last two
residues Ala263-Ile264, but the position of the C-terminus is quite
much varying from 1f4b and 1f4e.
SH 23.04.2007
