UHBD protocol
setting the grid
The
right size of the grid for UHBD calculations was determined by
superposition of sample structures of the relevant serine proteases and
align them to 1o5a. Used sample structures:
uPA
|
1lmw
|
thrombin
|
1a2c
|
factor Xa
|
1fax
|
trypsin
|
2ptn
|
plasmin
|
1bui
|
tPA
|
1a5h
|
final UHBD input files:
desolv1_35.inp
desolv2_35.inp
best
compiled UHBD version: /sw/mcm/app/uhbd/uhbd_linux_pp/uhbd110_PGFathlon
uhbd
read mol 1 file protein.qcd qcard end
read mol 2 file ligand.qcd qcard end
write grid binary file focus_bin.grd end
Comparing different grid sizes
desolv1_110.inp
Main stop
UHBD termination
date/time
: 14-Oct-05 19:07:13
total CPU seconds
: 522.760
total usr/sys CPU seconds :
516.290/ 6.470
desolv1_35.inp
large:
elec calc mol 1
pdie
2.00
! internal dielectric=2
sdie
78.00
! solvent dielectric=78
temp
298.00
! temperature in K
ions
150.00
! ionic strength
rion
1.5
! ionic radius
nmap
1.4
! use probe-accessible surface
nsph
400
! use 400 sphere points for surface cal.
bcfl
2
! boundary condition - each atom
! is a Debye-Huckel sphere
efld
.00
! external electric field
grid
1.00
! grid spacing
dime 65 81
71
! grid dimension
gcenter 15.0 -2.0 15.0
maxit
400
! maximum # of iteration for FDPBE
end
print elec phizero mol1
end ! zero
out the phisite accumulator
print elec phisave mol1
end !
compute and store phi at atoms on grid
focus:
elec calc mol 1
pdie
2.00
! internal dielectric=2
sdie
78.00
! solvent dielectric=78
temp
298.00
! temperature in K
ions
150.00
! ionic strength
rion
1.5
! ionic radius
nmap
1.4
! use probe-accessible surface
nsph
400
! use 400 sphere points for surface cal.
bcfl
4
! focusing - use the coarse grid to
! set the boundary potential of this
! focused grid.
efld
.00
! external electric field
grid
0.25
! spacing for focused grid
dime 109 109
109
! grid dimension
gcenter 9.0 5.0 26.0
maxit
400
! maximum # of iteration for FDPBE
end
print elec phisave mol1 end
print elec phinrg mol1
end !
compute and save to a user variable the
date/time
: 17-Nov-05 13:48:13
UHBD termination
date/time
: 17-Nov-05 13:59:58
total CPU
seconds : 293.030
total usr/sys CPU seconds
: 290.080/ 2.950
UHBD=/sw/mcm/app/uhbd/uhbd-6.1.expMSI_linux/bin.linux/uhbd_110_100000_65_wang
priamos
grid 109, 133.020
grid 100, 110.150
grid 109, !nmap !nsph, 133.370
grid 109, !nmap !nsph,ions=50, 136.470
epimetheus
grid 109, 142.830
prepare uhbd
uhbd
/sw/mcm/app/uhbd/uhbd_linux_pp/uhbd110_PGFathlon
step1.inp
read
mol 1 file A20.3.protein.qcd qcard end
read mol 2 file A20.3.ligand.qcd qcard end
! read in the coords., charges, and radii in CHARMM qcard
! format
!step 1
! Regular Runs:
!
compute the potential on a coarse grid for
!
molecule 1
elec calc mol 1
pdie
2.00
! internal dielectric=2
sdie
78.00
! solvent dielectric=78
temp
298.00
! temperature in K
ions
150.00
! ionic strength
rion
1.5
! ionic radius
bcfl
2
! boundary condition - each atom
! is a Debye-Huckel sphere
efld
.00
! external electric field
grid
1.00
! grid spacing
dime 81 81
81
! grid dimension
gcenter 9.0 5.0 26.0 !11
maxit
400
! maximum # of iteration for FDPBE
end
print elec phizero mol1
end ! zero
out the phisite accumulator
print elec phisave mol1
end !
compute and store phi at atoms on grid
!
compute the potential on a fine grid
!
molecule 1 using the coarse grid to set the
!
boundary potentials for this grid.
elec calc mol 1
pdie
2.00
! internal dielectric=2
sdie
78.00
! solvent dielectric=78
temp
298.00
! temperature in K
ions
150.00
! ionic strength
rion
1.5
! ionic radius
bcfl
4
! focusing - use the coarse grid to
efld
.00
! external electric field
grid
0.25
! spacing for focused grid
dime 109 109
109
! grid dimension
gcenter 9.0 5.0 26.0 !11
maxit
400
! maximum # of iteration for FDPBE
end
print elec phisave mol1 end
print elec phinrg mol1
end !
compute and save to a user variable the
! phi energy
assign e1 = $phinrg
end
! assign the phi energy to a local variable
! for later use
stop
priamos 95.960
epimetheus 67.980
/sw/mcm/app/uhbd/bin.linux/uhbd_110_100000_65_65_fast
step1.inp
calc_inter_model.sh
/sw/mcm/app/uhbd/uhbd_linux_pp/uhbd110_PGFathlon
step1-3: 269.010
step4: 266.230
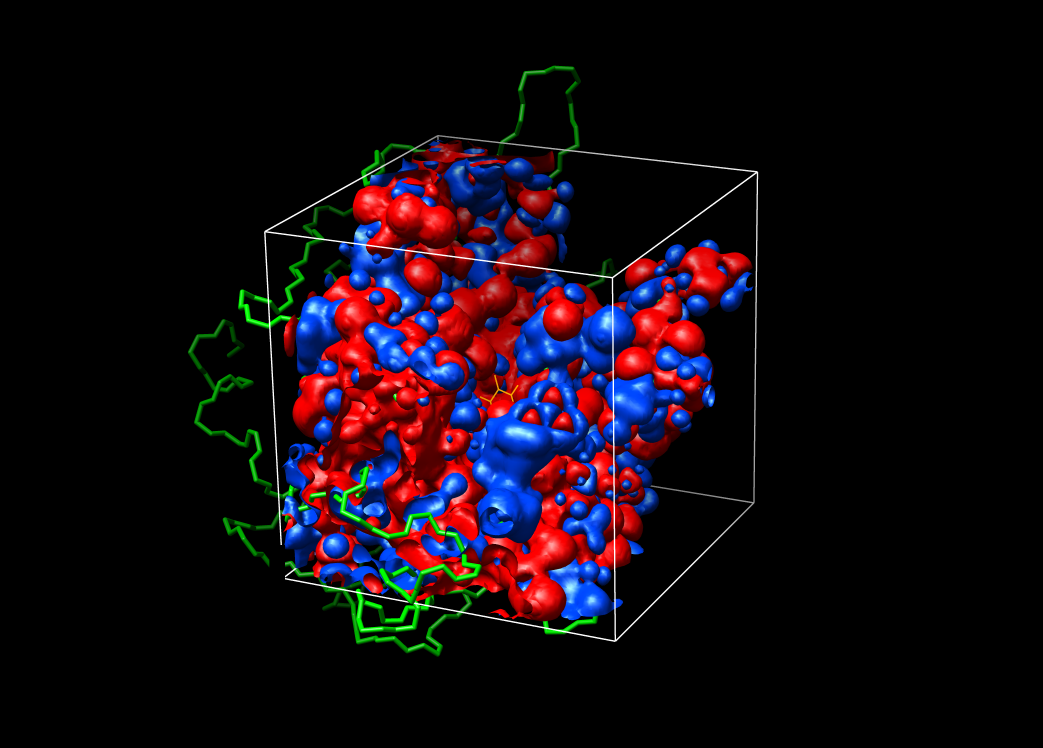
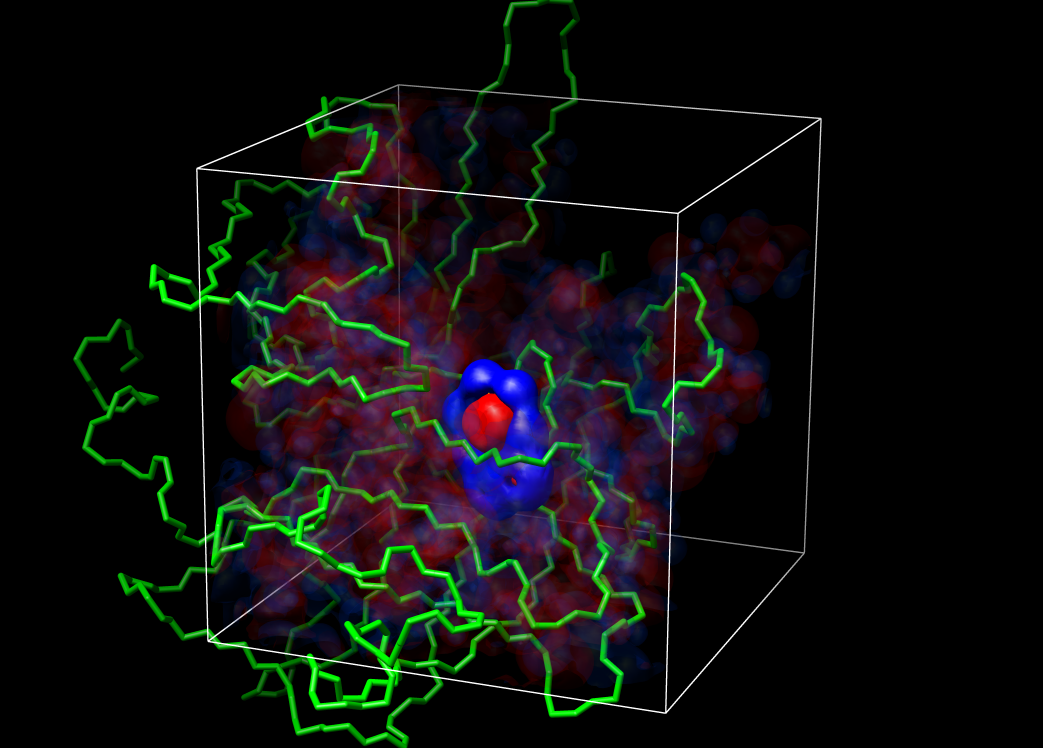
Visualization of the grid
in PyMol
pymol
converting grid:
/sw/mcm/app/vmd/aux/grid2i
grid2i B grid_uhbd.grd A grid_i.grd
draw x,y,z axis at origin
axes_cyl.py
Draw X, Y, and Z reference axes as CGO cylinders, colored red, green
and blue, respectively
load grid.grd, grid
isomesh msh, grid ,1.0,(model5),0.0,0
Loading binary UHBD grid, which
was converted by grid2i had wrong grid spacing and a shift in the
position!
Visualization of the grid
in chimera
chimera
prepare chimera
prepare chimera_121
chimera_send
Chimera was used for visualzation of the grids and electrostatic
potentials.
example grids:
/home/henricsn/combine2go/data/urokinase/prep-anal/model/model5_xray_medium/anal/
focus_109.grd large_81.grd
large grid (grid points 81, 81, 81; 1 A)
focus grid (grid points 109, 109, 109; 0.25 A)
surface of urokinase with benzamidine: blue=4, red=-4