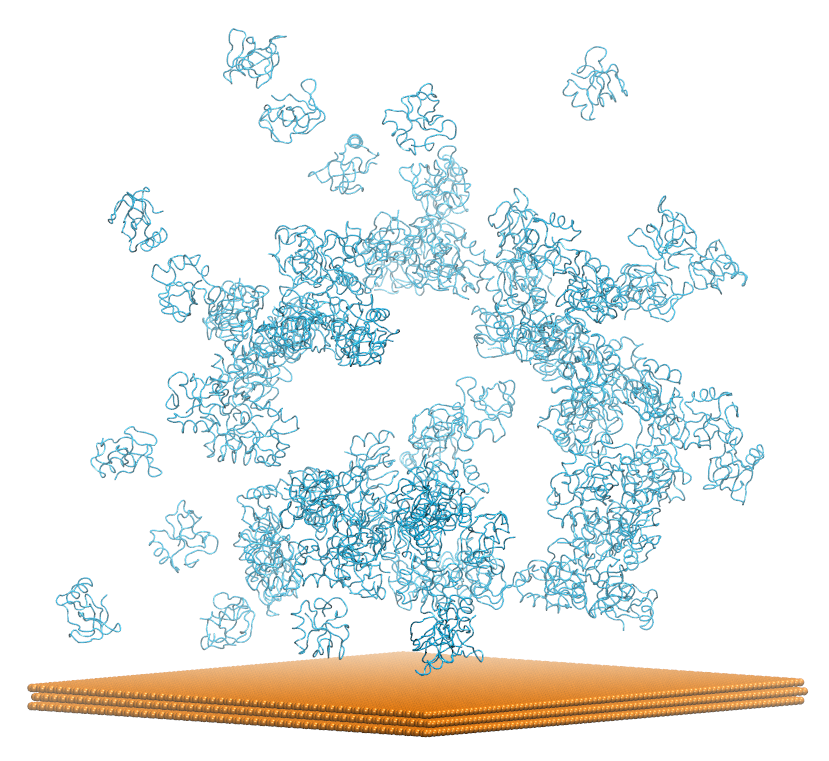
This example can be found in directory examples/HFBI-graphite.
You need to run at least one time the script in prepare_grids_and_ecm/ to generate the grids and effective charges needed by the input files.
| data_grid/ | Data files (pdb and input files) needed for running the examples |
| prepare_grids_and_ecm/ | Directory to prepare grids and effective charges |
| run/ | Directory to run the simulation |
| run/sdamm_out_hits | Subdirectory containing the ouput files generated at HITS. |
| doc/ | This documentation |
This input will run a sdamm simulation of 100 hydrophobin molecules with a graphite surface.
Graphite is a neutral surface, therefore the interactions between hydrophobins and graphite include only soft-core repulsion, electrostatic desolvation and non-polar desolvation.
However, the interactions among hydrophobins are represented fully, i.e., include the electrostatic term,
soft-core repulsion, electrostatic desolvation and non-polar desolvation.
To represent these asymmetric interactions correctly in SDA, one can omit the electrostatic grid and the effective charges for the graphite surface.
cd run/
./run_sdamm.sh
The script will run the simulation and generate a dcd format of the trajectory
You can compare the results with the output files in the subfolder: sdamm_out_hits/
A snapshot generated after the 25 ns of the multiple proteins run. It shows few interaction between the hydrophobins and the surface. The simulation should be run more than 100 ns to see spefic interactions.