This example (which is still in the testing phase) shows how to compute the dissociation rate constant for diffusional dissociation of barstar from a complex with barnase. For the input, it uses all pdb structures and grids precomputed in data_grid/. You need to run the script in prepare_grid_and_ecm/ at least once to generate the grids and effective charges in this folder.
cd bnbs_koff/
./script_koff.sh
This input will run a sda_koff computathe dissociation rate constant for diffusional dissociation of a solute from a complex. It is similar to a normal docking simulation, but the initial configuration should be read from a complexes file rather than starting randomly at a b-surface distance.
In addition, account can be taken of the occurrence of the configurations using the keyword account_occurency in the input file.
- If account_occurency is false (0), nrun trajectories are executed for every entry in the complexes file
- If account_occurency is true (1), nrun * occupancies trajectories are executed for every entry in the complexes file.
Occupancy is a specific field in the complexes file with the column header "Occur" (see complexes file for example).
It weights the complexes in the koff calculations.
If a restart file is not provided, the simulation will start from the absolute position of solute 2, taken from the pdb input file.
The option is similar to the SDA 6 implementation of sda_koff
sda_koff computes the average time that the reaction criteria are satisfied before unbinding (in the file residence_time), and
may be used to provide an estimation of k_off (results not validated).
The residence time is plotted on the figure below
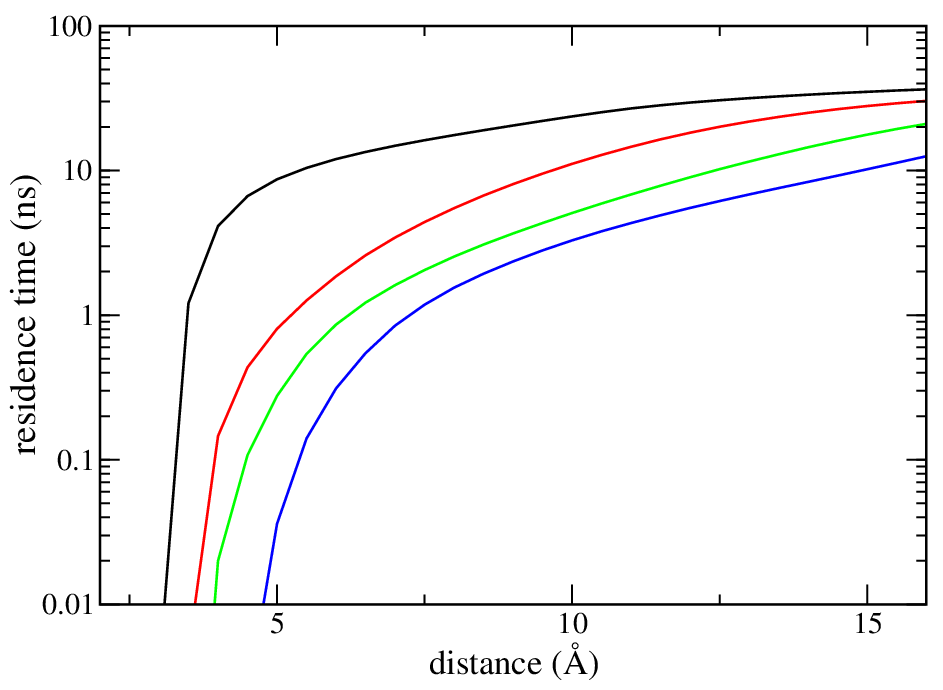
At 6 Å, for 2 contacts, it is approximately 1.8 ns.
It is more than was predicted by SDA 6. This option is still in the testing phase.


