This example can be found in directory examples/cytf_pc_et.
The grids and effective charges have been precomputed in the data_grid/ directory
| et_calc/ | Data files (pdb and input files) needed for running the Calculation of "Electron Transfer Couplings (T_DA)" and generation of "Reaction_Criteria_Files (.rxnac). |
| et_calc_scripts/ | Scripts required for the Calculation of "Electron Transfer Couplings (T_DA)" and generation of "Reaction_Criteria_Files (.rxnac). |
| data_grid/ | Data files (pdb and input files) needed for running the examples. |
| run_hits/ | Result of this example performed at HITS. |
| run/ | Directory to run the electron transfer simulation. |
| doc/ | This documentation and README file for Step-wise procedure need to be followed for Electron Transfer Rate Calculations. |
The setup of the simulation is similar to an association rate calculation. In the input file we must declare a ReactionCriteria and RateCalculation group, as for association rate.
In the calculation, it is the probability of electron transfer, dependent of the distance between the donor and acceptor pairs between the solutes, which is computed.
We used the coupling parameters given by the PATHWAYS software (A plugin for VMD).
The algorithm has been validated when using all 3 interactions:- Electrostatic
- Electrostatic desolvation
- Non-polar desolvation
Assure you have compiled all the executables and tools in sda_flex/bin/ first or refer to the compilation section.
Then execute the script in the run/ directory
cd run/
./run_cytf_pc_et.sh
You can compare the results with the ones provided in the run_hits/ direcotry
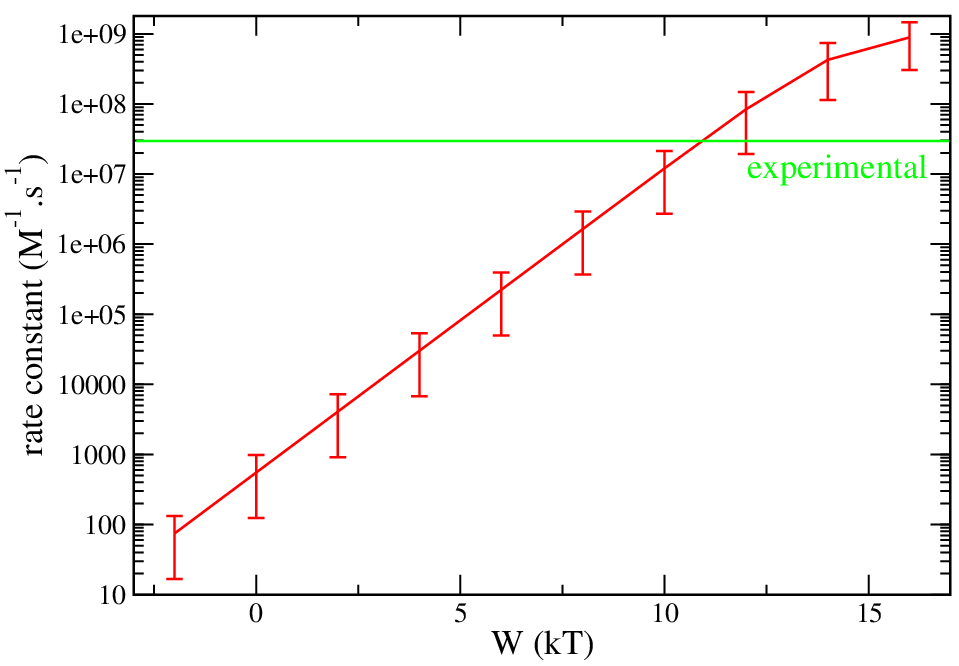
Electron transfer, cytochrome f and plastocyanin
parameters from the paper: W (in units of kT) of 10.4 reproduce experimental data (2.7E10+7 1/(M.s))