This script uses all pdb structures and grids in data_grid/. You need to run at least one time the script in prepare_grid_and_ecm/ to generate the grids and effective charges in this folder.
If you want more information about this step, refer to the tutorial in examples/Tutorial_SDA7/.
cd bnbs_assoc/
./script_assoc.sh
You can check that the output files are similar to the ones generated at HITS in bnbs_assoc_hits/
The script will run the calculation of the association rate between barnase and barstar in two different ways :
- 2 simulations with electrostatic and electrostatic desolvation terms:
- sda_association_bootstrap.in : run 1 longer simulation (2500 trajectories) with the
bootstrapoption set to 1. It creates the output file assoc_bootstrap.log which can be used to compute average and standard deviation with the python script auxi/Bootstrap_multiCPU.py - sda_association_X : runs five different simulations (with a different random generator, option
seed) of 500 trajectories. Average and standard deviation are computed with the auxi/nos2rates.f program. - 1 simulation of 2500 runs with the apolar desolvation term activated. Complexes and the first trajectory are printed and transformed into a dcd format.
Tips:
To draw the association rates you can use xmgrace : xmgrace -settype xydy rates_combined
To vizualise the film : vmd ../p2_noh.pdb trajectory1_apolar.dcd, and then load p1_noh.pdb (p1 never moves)
In SDA 7, rate constants are calculated using the Northrup, Allison and McCammon method (see Northrup, Allison McCammon, (1984), J. Chem. Phys.).
Simulations with only electrostatic terms reproduce the experimental association rate constant when the formation of 2 contacts (red curve) at ~6Å is used as the criterion for formation of an encounter complex.
Namely, three or four contacts are formed at the same rate with contact distances from 6 to 10 Å.
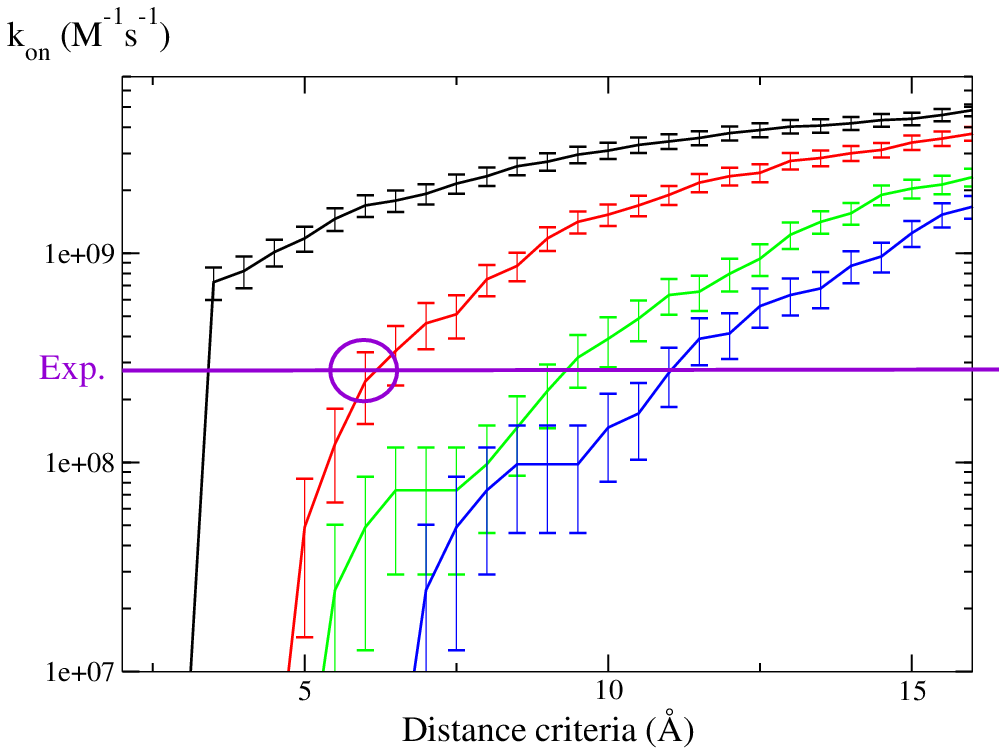
This graph has been drawn from the results in bnbs_assoc_hits/, from the simulation with bootstrap.
It is only 2 500 trajectories. Usually, we run 25 000-100 000 trajectories for producing reliable results.


