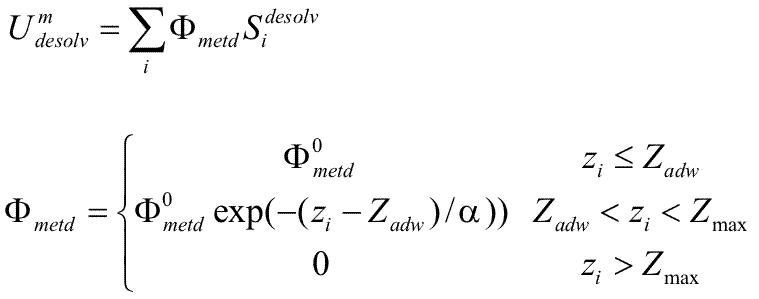SDA7 input file format
The input file consists of a series of grouped input
parameters, delimited by the keyword GROUP
GROUP = groupname
...
END GROUP
and a list of free parameters which are not associated with
a group.
All input parameters have the following form :
name-of-parameter
= parameter-value
Lines begining with the character "#" or "!" are treated as
comment lines and will not be parsed by the program.
Note: do not insert spaces in front of "#" if you want to
comment out a line!
Example input files
Detailed description of the input parameters
GROUP =
Type_Calculation:
type, total_solutes,
total_grids
-
type
[string]
(sda_2proteins)
: type of calculation -
defines which type of calculation will be performed.
The following options are available:
-
sda_2proteins
: for docking,
association rate calculation or electron
transfer
-
sdamm
: simulation with
multiple proteins.
-
sda_koff
: k_off
calculation.
-
sda_energy
: compute interaction
energy from complexes or trajectory files.
-
total_solutes [integer] (2) : total number of solutes
-
total_grids [integer] (2) : total number of grids (if a solute
has more than one grid to account for
conformational/structural variation, this only counts
as 1 grid ). In fact, the number of set_of_grids =
number of GROUP Solute_Grid
GROUP = Reaction
Criteria:
computation, rxna12f,
et_sol1, et_sol2, dind, nnnons, nwrec, sdamd
-
computation [string] (off_rc) : define which type of simulation is
being performed. Options:
-
association
: compute association
rate at every distance and for any number of
contacts defined in group RateCalculation. Use
*.rxna file and dind to define the criteria and
their independence, all criteria are considered as
nonspecific.
-
docking
: a complex must
satisfy all specific criteria and at least nnnons
non-specific ones in order to be saved in the
complexes file
-
electron_transfer
: compute elctron
transfer. The reaction criteria are read from 2
input files ( et_sol1 and et_sol2 )
-
all
: all complexes are
saved to the complexes file. No checks are
performed
-
off_rc
: disactivate the
reaction criteria module, no calculations
performed
-
rxna12f [string] (p12.rxna) : filename for the reaction criteria
used for docking or association rate
calculations.
Input examples for barnase-barstar docking
and association
rates are available in the examples.
Reaction atoms do not have to be real atoms, although
in this file these atoms/points should be given like
atoms in PDB format. Different combinations of
the same atoms/points can be prepared by editing these
reaction atom files
-
et_sol1, et_sol2 [string] (empty) : filenames for the reaction criteria
for electron transfer calculations.
-
dind
[float] (6.) : minimal distance (in Å) between atoms
on the same solute defining contacts for satisfying
reaction criteria (equivalent to d_min in figure
below)
-
nnnons [integer] (2) : minimum number of nonspecific
constraints to satisfy to define an encounter
complex
-
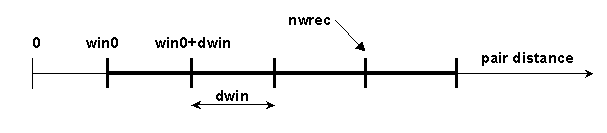
nwrec [integer] (0) : number of the window up to which the
complexes are recorded. For example, in the
situation below, nwin=5 reaction
distance windows are defined, and rates for forming
contacts at each of these 5 distances will be
monitored. Encounter complexes with contact
distances less than
win0+dwin*(nwrec-1), will be
recorded.
-
sdamd [integer] (0) : set equal to 1 to activate the option of
recording encounter complexes at the windows distance
according to win0+dwin*(nwrec-1).
For example, in the case where win0=4.0,
dwin=0.5, nwrec=23, encounter complexes will be
recorded with at least one reaction contact at a distance
of 15A. This option can be used in BD simulations of molecular association to
generate encounter complexes for subsequent molecular dynamics simulations.
Reaction criteria are used to define a "complex" between
2 solutes. Full freedom is let to the user:
- Define a center-to-center distance criteria between
the solutes, usually for docking
- A set of donor-acceptor pairs from an initial
complex, needed for association rates
- Any other constraints suitable with experimental
data: distance between 2 atoms satisfying crosslink
experiments, FRAP or FRET data, known interactions with a
specific residue / ligand...
- All constraints can be freely combined
The list of reaction criteria must be provided in a
separate file (conventionally named with a suffix
*.rxna), where the fields indicate:
- The keyword for the type of reaction criteria
(discussed below)
- The position of the atom of the solute 1 (the atom
number is needed in the case of a flexible solute)
- The minimum distance that the 2 reaction criteria
must satisfy
- The position of the atom of the solute 2 (the atom
number is needed in the case of a flexible solute)
For docking, there are 3 different types of reaction you
can use:
- CSPEC: specific constraint, this reaction criterion
must be satisfied for the complex to be accepted
- CNONS: nonspecific constraint, at least
nnnons of these reaction criteria must
be satisfied
- ASPEC: anti-specific, this criterion must be invalid
(new in SDA 7)
For association rate calculations, all entries in the
*.rxna file are considered as CNONS. But you can still
record complexes and adjust the nwrec
input parameter.
To get the specific format of the *.rxna file, we advise
you to look at the input files in the barnase-barstar
example of docking. Here are illustrated:
- A blind simulation, where only the center to
center distance between the protein is used as a
criterion
- An example where the center-to-center distance
criterion is used together with nonspecific reaction
criteria
Or you can refer to the technical documentation,
linked here in the
section Reaction Criteria.
(The doxygen documentation must be generated before using
this link).
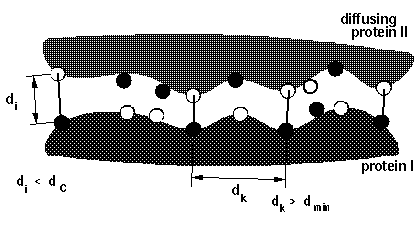
Usage of dind
To avoid an artificially large number of contacts, the
variable dind can be used.
On the figure below, all atoms used in the reaction
criteria are represented by spheres. But only those which
are at a separation greater than d_min are considered as
independent (black spheres). So with this schema, there
will be a maximum of 4 independent contacts.
GROUP =
RateCalculation:
win0, nwin, dwin,
nb_contact, bootstrap, fpt, stop_traj,analytical_correction
Calculate bimolecular association rate constants using the Northrup, Allison and McCammon method
(see
Northrup, Allison McCammon, (1984), J. Chem. Phys.).
-
win0
[float] (3.0) : reaction distance window minimum
value (in Å)
-
nwin
[integer]
(0) : number of reaction distance
windows.
-
dwin
[float] (0.5) : reaction distance window step (in
Å).
-
-
nb_contact [integer] (4) : maximum number of pair contacts to be
considered. This parameter is only applicable for
association rate calculations.
-
bootstrap [bool] (0) : if 1, association rates and first
passage time results are printed for every trajectory.
Final results can be computed with bootstraping
(auxi/Bootstrap_multiCPU.py)
-
fpt
[bool] (0) if 1, record the first passage time, if
bootstrap==1 same processing with
(auxi/bootstrap.py)
-
stop_traj
[bool] (0) if 1, simulated trajectories will stop when the most
strict reaction criteria definition is met (i.e. greatest number of contacts
at the closest distance window). This may speed up your simulations, particularly
in the case of strongly interacting simulations, although this will depend strongly
on your choices of nb_contact and win0.
-
analytical_correction
[bool] (0) if 1, the analytical correction for centrosymmetric forces is computed for estimating the bimolecular association rate constant with the Northrup, Allison and McCammon method
. When using this option, it is required that
the interaction potential is centrosymmetric at the b surface, but it can be non-zero. This option requires
that the real_net_charge and dh_radius is set for all solutes in the
solute grid groups of the input file, and that the ionic_strength is set in the analytical
group. If 0, there must be no interaction forces between the solutes at a separation of start_pos
GROUP =
Analytical:
h_analytic, debyeh, ionic_strength, hydrodynamic, disable_normal_hi,
disable_image_hi, disable_height_hi, surface_height, hom_charged_surf, surface_fact, gouy_chapman
-
h_analytic
[float] (0.01) : defines the bin size (in Å) of the arrays of the precomputed debye-hueckel values.
-
debyeh
[bool] (0) : if 1, activates Debye-Hueckel interactions for all solutes
-
dh_imgchg
[bool] (0) : if 1 and a surface is present, the image-charge model used for
modelling electrostatic interactions with conducting surfaces is extended to use
debye-huckel interactions for all solutes whose images lie outside of their
electrostatic grids.
-
ionic_strength
[float] (0.150) : if Debye-Hueckel computed, specifies the ionic strength (in Molar)
-
hydrodynamic
[bool] (0) : if 1, activates mean-field hydrodynamics interactions for all solutes. If a surface
is present in the simulation then surface effects will be included. This can be disabled
with the options disable_normal_hi, disable_image_hi
and disable_height_hi. These options are meant for investigating the effects of
hydrodynamic interactions of various types, and for debugging. In most cases all hydrodynamic
components should be included.
-
disable_normal_hi
[bool] (0) : if a surface is present and this option is set to 1, normal mean-field
hydrodynamics interactions between solutes are disabled.
-
disable_image_hi
[bool] (0) : if a surface is present and this option is set to 1, mean-field
hydrodynamics interactions between solutes and their surface reflected images are disabled.
-
disable_height_hi
[bool] (0) : if a surface is present and this option is set to 1, the height dependent
slowdown in diffusion close to the surface is disabled.
-
lvol_rcut
[float] (0.0) : defines the local volume radius if hydrodynamics interactions are used. Must be greater than the largest vol_radius of any solute in the system.
-
surface_height
[float]
(0.0)
: defines the height in Angstroms above a surface at which the image method is applied for surface hydrodynamic interactions. Typical value used in the Lysozyme_mica example is 2.0.
-
hom_charged_surf
[bool]
(0)
: if 1, the long-range Debye-Hückel electrostatic treatment for a surface is activated
-
surface_fact
[float]
(0.0)
: defines the prefactor for the surface charge potential (1.0 = (infinitely) thick surface (default for the Lysozyme_mica example); 0.5 = thin surface, e.g. represented by one atom layer
-
gouy_chapman
[bool]
(0)
: if 1, use Gouy-Chapman treatment instead of Debye-Huckel for the long range electrostatics of the Surface. The Gouy-Chapman treatment is the solution to the non-linearized
Poisson-Boltzmann equation for a wall-geometry, whereas the Debye-Huckel treatment models the linearized Poisson-Boltzmann equation.
-
surface_fact
[float]
(0.0)
: defines the prefactor for the surface charge potential (1.0 = (infinitely) thick surface (default for the Lysozyme_mica example); 0.5 = thin surface, e.g. represented by one atom layer
GROUP =
Solute_Grid:
nb_solute, pdb_filename, sasa_filename,
diffusion_trans, diffusion_rotat, real_net_charge, surface_charge_dens, rotate, surface, flex,
epf, qef, edf, hdf, ljf, lj_repf, nb_lj, atom_lj,
list_conformation, dh_radius, vol_radius, total_conf, method,
initial_conf, frequency, std_frequency, surface_charge_dens
-
nb_solute [integer] (1) : number of identical solutes ( of the
same type)
-
pdb_filename [string] (empty) : name of pdb file for this group of
solutes
-
sasa_filename [string] (empty) : to calculate non-polar desolvation forces using
customized solvent accessible surface area (sasa) values, provide the name
of the file (pdb format) containing said sasa values. If sasa values in
pdb format are not provided, they will be read from *.access file.
-
diffusion_trans [float] (0.0123) : absolute translational diffusion
coefficient in Å2/ps. In the case of
sda_2proteins, the translational diffusion constant of
solute 2 should be assigned as the sum of the diffusion
coefficients of solute 1 + solute 2 to simulate the
relative translational diffusion.
According to the Einstein-Stokes formula, the
translational diffusion constant scales inverse
proportionally to the solvent viscosity (which itself
has a complicated dependence on the temperature), and
inverse proportionally to the (hydrodynamic) radius of
a solute. 1 Å2/ps = 10-8
m2/s = 10-4
cm2/s
-
diffusion_rotat [float] (1.36e-4) : rotational diffusion coefficient of
the solute in radian2/ps. This is inversely
proportional to the square of the solute radius.
diffusional_rotat is inversely
proportional to the solvent viscosity. For lysozyme,
for example, dr=2.6*10-5
radian2/ps.
-
real_net_charge [float] 0.0 : Total net formal charge on the solute.
Note that this is generally different to the sum of the
effective charges which are used in calculating electrostatic
forces and energies via interaction with Poisson-Boltzmann derived
potential grids.
The charges set here are used to approximate long range charge interactions between
solutes using a Debye-Hueckel sphere approximation, if debye is
set in the Analytical group, and in the analytical correction to the rate constant
calculations, if analytical_correction is set in the RateCalculation
group.
-
surface_charge_dens [float] (0.0) : If the solute is a surface, this option can be used to define an
average charge density for surface. The interaction with other solutes is then modelled
using a Debye-Hueckel sphere approximation where spherical charge solutes interact with
a homogeneously charged surface. If an electrostatic grid is included in this
Solute_Grid group, then the approximatation is made in regions outside the grid.
With this option ionic_strength must be set in the Analytical group.
Unit is in e/Angstrom**2
-
rotate [bool] (1) : switch to define whether the solute
rotates. 0 means no rotation and
diffusion_rotat is not used. 1 means
the solute can rotate and
diffusion_rotat is used.
-
surface [bool] (0) : switch to define whether this solute
is a surface. Being treated as a surface will influence
the distance computation and periodic boundary
conditions, and rotate will be set to
0 for this solute
-
vert_excl [bool] (0) : if set to 1 in the Solute_grid group of
the central solute in a bimolecular simulation, the minimum
solute separation at which it is assumed safe to ignore checks
for excluded volume violations depends only the
z coordinate of mobile solute, rather than the centre-to-centre
distance. This is always assumed if the central solute is
defined as a surface.
-
xcent, ycent, zcent [float] (geometric centre of solute) : options to set the centre point of the central
solute in a bimolecular simulation. The coordinates should be defined
in the coordinate frame of the input PDB file. If not present in the input file
then the geometric centre of the solute is used. This centre point is
used to define the location of the starting and ending points of the
trajectories (defined by start_pos and c
in the geometry group. This can be useful in the case of a surface, or
otherwise non-spherical solute, as it allows the simulation volume to
be centred on a surface lying point. If one of these variables is set, then
all must be.
-
flex
[bool] (0) : define if this solute(s) is flexible
(i.e. has more than 1 structure/conformation) or
not
-
image_charge [bool] (0) : define if the electrostatic image
charge of this solute(s) must be computed. It is
relevant in the case that one solute is a metal
surface.
-
stoke_radius [float] (0.) : Obsolete: This option is now replaced by the radii options
shown below.
-
dh_radius [float] (0.) : if > 0, indicates the radius used to represent the solute
cavity in the Debye-Hueckel sphere charge model.
-
This option can be used as a long range correction to the truncation of electrostatic
grids in all-atom simulations, and in coarse-grained sphere simulations
and adaptive-resolution crowder simulations.
-
vol_radius [float] (0.) : if > 0, indicates the radius used
for calculating the local occupied volume fractions of the mean-field hydrodynamic interaction model.
-
Hydrodynamic interactions are implemented with
the Weinstein method, where the diffusion coefficients
are rescaled during the trajectory depending on the
instantaneous local volume fraction of solutes around
one solute. The infinite dilution values of the
diffusion coefficients can be obtained from the
HYDROPRO program, for instance, or determined from
experiments. See Mereghetti et al. (2012)in
references
-
epf
[string]
(empty)
: filename for the
electrostatic potential of the solute in UHBD binary
format. It is assumed that the potentials are
written in kcal/mol/e units. Note that APBS
generated grids written in UHBD format are in units of
kT/e and ascii and as such cannot be used directly with
SDA. They should be rescaled and converted with
the auxiliary program convert_grid, to bring them
into consistency with UHBD format.
-
qef
[string]
(empty)
: filename for effective
charges of the solute. This is the output of the
ECM suite program. See
detailed description in the faq
-
edf
[string]
(empty)
: filename for the
electrostatic desolvation potential of the solute in
UHBD format. This grid can be calculated with the
program make_epedhdlj_grd
which is included in the SDA distribution (units in
kcal/mol/e).
-
hdf
[string]
(empty)
: filename for the
hydrophobic desolvation potential of the solute in UHBD
format. This grid can be calculated with the program
make_epedhdlj_grd which is
included in the SDA distribution.
-
lj_repf [string] (empty) : filename for the soft-core repulsion
(approx. repulsive term of Lennard-Jones) grid of the
solute in UHBD format. It is intended to be used with
sdamm only where it replaces the use of the exclusion
grid. This grid can be calculated with the program
make_epedhdlj_grd which is
included in the SDA distribution.
-
nb_lj [integer] (0) : define the number of Lennard-Jones
potential grids. Used for the implementation of the
ProMetCS force field.
-
ljf
[array of string]
(empty)
: filenames for the
Lennard-Jones potential grids of the solute in UHBD
format. This grid needs an additional tool to be
computed
-
atom_lj [array of string] (empty) : atom names for the Lennard-Jones
interactions. If an unusual atom name, use the
"add_atoms" file to define the sites of
interaction.
-
These variables are only needed in the case where
the flag flex is set to 1
-
list_conformation
[string]
(empty)
: filename of the file
which defines multiple conformations/structures for a
solute. Every GROUP SoluteGrid may have a different
list.
-
total_conf [integer] (0) : define the total number of
conformations/structures, if this solute is treated as
flexible
-
method [string] (empty) : choose between the methods
implemented for changing conformations/structures.
Available options are:
-
-
random
: Choose a random
conformation (depending on nearest
input). The same conformation can be chosen.
Energies are not evaluated
-
minimum
: Implemented only with
nearest=1. The minimum energy
between the actual and the closest conformations (1
or 2 ) is selected
-
metropolis
: The potential energy
in the initial conformation (E0) is computed. For a
random chosen conformation ( depending on
nearest ), the new energy (E1) is
computed and the move is accepted if:
-
-
the new
conformation has a lower energy: E1 is less
than E0, or
-
a random number
uniformly generated in the range [0,1] is lower
than exp(E1-E0)
-
nearest [bool] (0) : If 1, moves are limited to the
nearest neighbour ( conformation +/- 1). It is used
with conformations from normal mode analysis for
instance.
Otherwise a random conformation is chosen (used with
NMR structures). The same conformation can be
selected.
-
initial_conf [integer] (1) : Specify the conformation chosen at
the beginning of every trajectory.
-1 makes a random choice
-
frequency [float] (100) : Specify the delay between the changes
in conformation (in ps). The delay is computed as a
Gaussian random number with average
frequency and standard deviation
std_frequency.
-
std_frequency [float] (0) : Specify the standard deviation (in
ps). Used in the case of SDAMM to avoid synchronisation
between the solutes.
GROUP =
Geometry:
type, pbc, surface, escape,
start_position, c_surface, min_X, max_X
-
type [string] (sphere) : specify the geometry of the
simulation box
-
sphere :
: Use a spherical geometry,
only for use with sda_2proteins types of
simulation
-
box :
: Use a box for simulation,
only for use with sda_2proteins types of simulation
(protein-surface case) and with sdamm
-
pbc
[bool] (0) : Switch on Periodic Boundary
Conditions. Apply only with a box geometry
-
surface [bool] (0) : Indicates if a surface is present in
the system. PBC rules are modified in this case. Can
also be used with a spherical geometry in sda_2proteins
simulations
-
escape [bool] (1) : Indicates if a solute can escape. The
trajectory is then stopped when the solute reaches the
c_surface (with a sphere) or zmax (in a box)
-
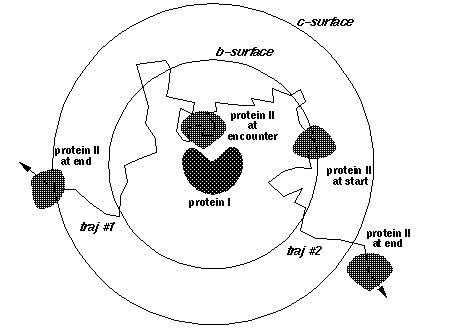
start_pos [float] (100.) : Indicates the initial position (in Å)
of the second solute in sda_2proteins. Simulations are
started by placing the center of the second solute at
the distance start_pos .
This corresponds to the b-surface for a spherical
geometry.
If surface is activated, the initial
position is generated on the upper-half of the
sphere.
For calculation of association constants, there should
be no interaction between the solutes at this
distance if analytical_correction = 0, or the interaction should be centrosymmetric if analytical_correction = 1. To ensure there are no grid-based interactions at this distance,
start_position can be selected to be
larger than the sum of the solute 1 (or 2) grid extents
and the solute 2 (or 1) radius.
To check with box.
-
c
[float] (150.) : defines the c-surface (in Å) in the
case of a sphere
-
<X>min,<X>max
[float] (0.) : where <X> = x, y or z.
Defines the size of the box (in Å). If
escape is activated, zmax has the role
of the c-surface.
-
half_sphere
[bool] (0) : Replace the spherical b surface with a
spherical cap surface above an xy plane at a height z =
min_height. This option is always used
in the presence of a surface.
-
min_height
[float] (0.0) : defines the height of the spherical cap,
relative to the centre of the centre solute,
when option half_sphere is used.
Typical setup for simulations within a sphere.
Trajectories are started at the b-surface
(start_pos) and finish when solute 2
goes through the c-surface (c).
GROUP =
Timestep:
variable,
dt1,
swd1,
dt2,
swd2
-
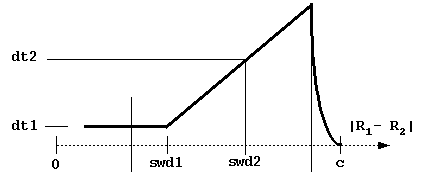
variable [bool] (1) : switch to a variable timestep ( in
sda_2proteins simulation), or to a fixed timestep
(sdamm) when the value of dt1 is
used
-
dt1
[float] (1.0) : basal simulation timestep (in ps,
picoseconds). This is the smallest timestep used
and is employed when the center-to-center separation is
less than swd1 or the distance at
which the solutes have the possibility to contact (i.e.
the center-to-center distance between solutes is less
than rhit = the sum of maximal extents
+ probe radius + maximal radius of solute 1
atoms).
-
swd1
[float] (50.0) : center-to-center distance (Å) at
which the timestep starts to increase
-
dt2
[float] (20.0) : timestep at distance
swd2
-
swd2
[float] (90.0) : center-to-center distance at which
timestep equals dt2.
-
height_dependent_dt
[bool] (0) : if set to 1, the variable timestep
depends on the z coordiate of the mobile solute, rather
than the centre-to-centre distance between the solutes.
This option is always used when a surface is present.
-
rswd
[*] (*) : deprecated in SDA 7, the rotation of
solute2 is only proportional to the timestep
These parameters define the variable timestep designed to
accelerate simulations. As shown in the figure below, the
timestep is automatically decreased to zero near the
c-surface to reduce artifacts due to truncation of the
trajectories. Although the profile of the time step
influences the computed rates, the most important
parameter is dt1. Take care not to choose a too small
value for this parameter (since the computing time scales
almost linearly with it). The physical parameter
sqrt(dt*dm) measures an average Brownian dynamics step
length. This must be less than 1 Å, which is the
characteristic distance describing the roughness of any
protein surface since the atomic bond lengths and van der
Waals radii define this scale. Moreover, dt1 should be
less than the characteristic distance over which the
interaction forces vary substantially, which means that
the optimal value is influenced by interaction force
strength: one needs smaller time steps for stronger
interacting proteins. The standard output will print
additional information, like the location and the value
of the maximum timestep.
GROUP
= ResidenceTime:
type_residence,
fixed_bin_size, size_X,
cell_X,shift_rt_X,format_rt3d,pair_solutes
-
type_residence [string] (off) : can use different geometry
-
-
off
: default, disactivate
the module
-
distance
: record the
center-to-center distance between the
solutes
-
plan
: project on a
surface
-
resid_3d
: record on a 3d
grid
-
fixed_bin_size [integer: 0,1,2] (0) : adjust the size of the
arrays/grids
-
-
0
: the number of cells
is fixed (cell_X), adapt the size to fill up to the
c-surface
-
1
: fix the size of one
cell (size_x), adapt the size to fill up to the
c-surface
-
2
: fix both the number
of cells (cell_X) and the size of one cell (size_X)
to allow to focus on the solute 1
-
size_x, size_y, size_z [float] (1.0) : size of each cell (in Å), the UHBD
format allows only an identical spacing for
visualisation
-
cell_x, cell_y, cell_z [float] (20.0) : number of cells in each
direction
-
format_rt3d [bool] (0) : Switch to choose between ascii or binary output format. Default is binary.
-
pair_solutes [bool] (0) : Switch on to compute rdf for all pairs of solutes in multiple molecule simulations. Default is just one pair as required for two-solute computations.
GROUP =
Complexes:
fcomplexes,
restart_complex,
binary_complex,
nb_complexes,
rmsd_min,
ionerun,
complex_sum_energy
merge_step,
size_thread,
ftrajectories,
binary_trajectory,
ntraj_rec,
freq_print,
trajectory_sum_energy,
max_array_traj,
mem_traj_frames,
max_array_traj
-
fcomplexes [string] (empty) : filename to record output complexes.
By default this is switched off, no complexes are
recorded.
-
restart_complex [bool] (0) : If switched on, the complex file will
be first read during the initialisation. This is used
in the case that we want to continue a simulation. Note
that it is important to change the random number
generator seed, otherwise you may reproduce identical
trajectories.
-
binary_complex [bool] (0) : Switch to choose between ascii or
binary output format. Ascii files are human readable,
but binary improves the precision of the numbers.
binary may also save disk space, but this is relatively
limited. Default is ascii.
-
nb_complexes [integer] (500) : No. of distinct lowest energy
configurations to write. Complexes with a higher energy
are discarded
-
rmsd_min [float] (2.0) : minimum rmsd between complexes in Å.
During the BD simulations, if a new docking pose is
considered similar to a previously stored pose, then
the configuration with the lowest total energy is
stored and the counter for this docking pose is
increased. A pose is considered similar to a previous
pose if they have an RMSD less than the designated
threshold
-
ionerun [bool] (1) : Switch to choose whether rmsd values
are compared with other trajectories if ionerun = 0,
or the default of only comparing within an individual trajectory if
ionerun = 1. It is usually advisable to change this to ionerun = 0.
-
complex_sum_energy
[bool] (0) : Switch to choose the format of the
energy terms. If set to zero ( default ) the energy
components are split into the energy of the solute 1 in
the grid of the solute 2 and its inverse. If set to 1,
only the sum is printed out. We expect the
electrostatic energy terms to be similar. Due to the
approximation in the PB calculations, grids and
effective charges the energies cannot be exactly the
same, but should be approximately the same. If a
difference of a factor of more than 2 occurs, this may
indicate an inadequate electrostatic and/or effective
charge calculation.
-
merge_step [integer] (input dependent)
: In case of OpenMP only,
indicate how often every thread merges their complexes
into a global complexes file. The default values are
chosen from different parameters in the input
file.
-
size_thread [integer] (input dependent)
: In case of OpenMP only,
indicate the size of the complexes list of each
thread.
-
ftrajectories [string] (empty) : filename to record the trajectory
output. By default this is switched off and no
trajectory is recorded.
-
binary_trajectory [bool] (0) : Switch to choose between ascii or
binary output format. Ascii files are human readable,
but binary improves the precision of the numbers. The
effect on the number precision is clearly seen in the
case of a sdamm trajectory. If set to 1, the restart
file will also be saved in binary format. Binary may
also save disk space, but this is relatively limited.
Default is ascii.
-
ntraj_rec [integer] (-1) : trajectory no. to be recorded. If the
value is negative, all trajectories will be recorded.
If the value = 0, no trajectories will be recorded. If
the value is positive, record only this trajectory
number.
-
freq_print [integer] (100) : frequency ( in steps ) with which the
conformations are recorded in the trajectory
file.
-
trajectory_sum_energy
[bool] (1) : Like complex_sum_energy
for the trajectory. The algorithm is written to apply
with any number of energy components. Default is that the sum is printed out.
-
max_array_traj [integer] (nb. of solutes) : DEPRECATED: Maximum size of the array for the
trajectory. The array is printed to file when
filled.
-
mem_traj_frames [integer] (1) : Number of trajectory frames to store in
memory before writing to disk. If trajectory frames are written
frequently, increasing this parameter could increase simulation
performance when it is limited by frequent disk access. The replaces
the now deprecated parameter max_array_traj.
GROUP =
MetalDesolvation:
distance_to_surface,
energy_per_water, radius_patch, radius_water
- This energy term has been
parametrized for computing Au(111)-protein interaction
energies. Computation of forces has not been
parameterized:
-
distance_to_surface
[float] (6.5) : in Å (Zadw); the position of the
first hydration layer (3 Å) plus the average LJ radius
of protein-surface interaction
-
energy_per_water [float] (0.13) : Φ°_metd in kT/Ų (Eadw); desolvation energy
per unit area of the first hydration
layer
-
radius_patch [float] (6.0) : α in Å; (Aadw) parameter for
calculation of the surface desolvation potential Φ as
parametrized from MD simulations of PMF for the test
atom on the Au(111) surface
-
radius_water [float] (1.5) : Radw in Å; The effective desolvation
radius that defines the surface area that each protein atom
is assumed to desolvate.
The surface desolvation energy is calculated as :
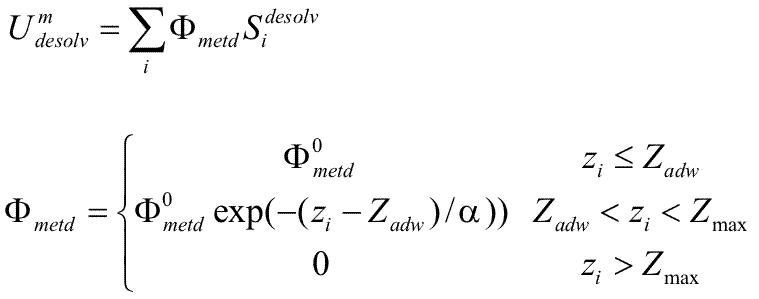
The computed desolvated area is shown by bold lines, the
solid and dashed lines corresponding to water desorption
from the first and second hydration layers, respectively.
The circles with crosses represent close solute atoms and
that with a zero is at an intermediate distance.
GROUP =
PMF:
mentth, mentfi, mentom, mentx, menty, pstart,
pfinish, dz
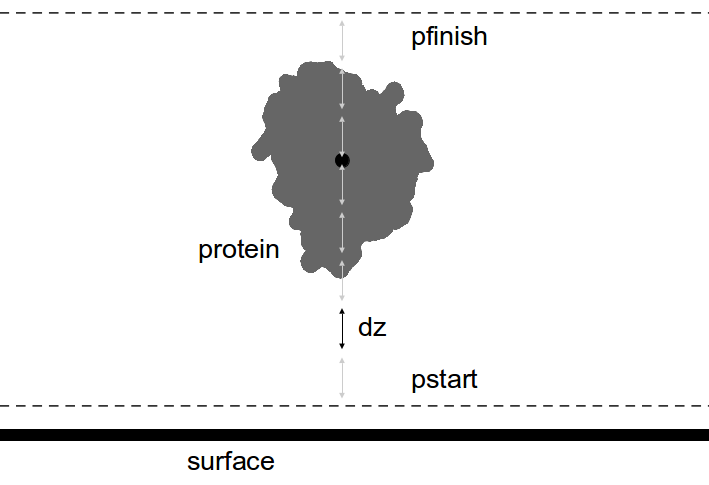
- Potential of mean force (PMF) calculations
are based on a thermodynamic integration method where
the configurational space of the solute with respect to a surface is sampled over the Euler angles
(θ, ϕ, ω) and a number
of dx,dy and dz coordinates based on the parameters:
-
mentth
[integer] (60) : Number of θ angles to be explored in rotational sampling.
The angles range between 0 and π with a stepwise increment of (π ÷ (mentth - 1))
-
mentfi
[integer] (120) : Number of ϕ angles to be explored in rotational sampling.
The angles range between 0 and 2π with a stepwise increment of (2π ÷ (mentfi - 1))
-
mentom
[integer] (60) : Number of ω angles to be explored in rotational sampling.
The angles range between 0 and 2π with a stepwise increment of (2π ÷ (mentom - 1))
-
mentx
[integer] (6) : Number of subdivisions (dx) of the surface along the x axis to be sampled.
The sampling length in the x direction is 6 Å
-
menty
[integer] (6) : Number of subdivisions (dy) of the surface along the y axis to be sampled.
The sampling length in the y direction is 6 Å
-
pstart
[float] (0.0) : Starting distance of the center of geometry of the solute from the surface in Å
-
pfinish
[float] (60.0) : Finishing distance of the center of geometry of the solute from the surface in Å
-
dz
[float] (0.2) : Increment step (dZ) along the z axis in Å
Parameters not associated with a group, they apply to all
solutes
-
dseed [float] (256.) : random number generator seed. If the
value is 0, it uses the time clock. With OpenMP, it is
not guaranteed that the same trajectory will be
obtained with the same seed.
-
nrun
[integer]
(1) : number of trajectories to generate.
Only 1 is possible with SDAMM. The higher this number,
the higher is the number of reactive trajectories
(nrun_reactive). The relative error of the calculated
rate constant is ~ 1/sqrt(nrun_reactive). This means
that to have the same relative error, a larger value of
nrun is required in cases with lower association rate
constants.
-
timemax [float] (0.0) : maximum length of computed
trajectories in ps.
If timemax = 0.0, there is no limit; this is the recommended value for
two-solute association rate calculations.
-
old_orientation [bool] (0) :Turn on the old version of computing the rotation matrix from the torque vector.
In the new implementation, this conversion is performed using Rodrigues formula. However, in the old implementation,
this conversion is performed by carrying out rotations first around the z-axis, then the y-axis and then the x-axis. This only works
for infinitesimal rotations up to first order.
-
nprint : deprecated; frequency with
which rate of information is printed: the fraction of
reactive trajectories is printed every nprint
runs. This is most useful when the simulation
takes a long time, then intermediate results are
available after each nprint trajectories.
Values of the different probes
-
probep [float] (1.7) : radius of the probe used to compute the exclusion volume grid.
The surface of solute 1 that is excluded to solute 2 is
defined by increasing the radii of the atoms of solute 1 that have a solvent
accessibility greater than threshold
by
probep (Å). probep is usually chosen to have a value corresponding to the average radius of the surface atoms of solute 2, e.g. 1.77 Å for a protein. Typical values of this
parameter are 1.4-1.9 Å. A smaller value (0.5 Å) should be used with the ProMetCS force field as it includes a Lennard-Jones term.
-
probew [float] (1.4) : radius of solvent probe used to
calculate the solvent accessibilities of solute atoms. A
recommended value to use is 1.4 Å to represent a water molecule. This value is
especially important if the hydrophobic desolvation
term is used, because the calculated solvent accessibilities
are used to calculate the hydrophobic desolvation
energy and forces.
-
threshold [float] (0.0) : solvent accessibility threshold - only the
atoms of solutes with solvent accessible area greater than
threshold (Ų) are designated as
surface atoms. Moreover, only these atoms are used in
calculating hydrophobic interactions and soft-core
repulsion (repulsive Lennard-Jones). The default value
is 0.0. This value is recommended for calculations with two solutes. A value of 5.0 is recommended for multiple solute (sdamm) simulations and for these, genbox, rather than SDA, should be used to compute the solvent accessibilities. The threshold value of 5.0 is used by genbox (with probew = 1.4 Å) to compute the solvent accessibility file for each solute. These files are used by SDA if save_access is set to 1.
-
hexcl [float] (0.5) : spacing of the exclusion grid with
which a solute is represented. Does not apply to SDAMM
calculations where the exclusion grid is replaced by a
soft-core repulsion. hexcl defines the
accuracy with which the shape of the solute is
described. Typical values are 0.5-1.0 Å for all-atom
models of proteins. It needs to be consistent
with the timestep used for BD moves - the spatial
accuracy of the protein representation should be
comparable to an average BD move in realistic
simulations.
-
Saving data computed during the initialisation phase
The files will be saved in the directory where the
pdb files are deposited.
save_exclusion [integer,0,1,2] (0) : if > 0, tries to read (and write)
the exclusion grid from/into the disk. The grid can be
loaded in VMD.
-
0
: force recomputation
of the exclusion grid during the
initialisation
-
1
: save the grid in
binary format
-
2
: save the grid in
ascii format
save_access [logical] (0) : if 1, tries to read (and write) the
list of accessible atoms from/to disk. It may save a
lot of time if there are more than 100 000 atoms
present in a pdb file. It is recommended to set save_access to 1 for multiple molecule (sdamm) simulations with solvent accessibility files generated by genbox.
-
0
: force recomputation
of the accessible atoms during the
initialisation
-
1
: save the data in
ascii format
Multiplicative factors applied to the grids
SDA allows simulations with intersolute forces computed from different combinations of contributing terms that should be set by the user.
- ---For simulations of two solutes with excluded volumes and no intersolute forces, set all the following factors to zero. For simulations of multiple solutes that do not interact except through soft core repulsion, set all the following factors to zero except lj_rep_fct which should be set to 1.0.
- ---Set ljfct=1.0 if simulating a protein diffusing to a metal surface with the ProMetCS force field. Otherwise, ljfct should be set to 0.0.
- ---For simulating a single protein or peptide diffusing to a metal surface with the ProMetCS force field, all factors except lj_rep_fct should be set to non-zero values. Set epfct=0.5, edfct=3.34, hdfct=-0.0065, ljfct=1.0, see below.
- ---To compute the electrostatic interactions beyond the extent of the electrostatic potential grid of a solute molecule using the Debye-Hueckel model, set debyeh=1 (see GROUP = Analytical). This is advisable for calculations at low ionic strength or if the size of the electrostatic potential grid needs to be restricted.
- ---For computing bimolecular association rate constants for two proteins, set epfct=0.5 and set edfct and iostr in ed.in as described below. The other factors are usually set to 0.0.
- ---For docking two proteins to generate diffusional encounter complexes, set epfct=0.5 and set all other factors to 0.0. This setting was used for rigid-body docking subject to biochemical constraints by Motiejunas et al. Proteins, 2008, 71:1955-1969
Article
link. The encounter complexes generated were refined by short molecular dynamics simulations, allowing for molecular flexibility, and the procedure was evaluated for a diverse set of protein pairs. This setting, using the electrostatic interaction term only, has also been used for rigid-body docking of proteins to solutes containing nucleic acids. An alternative that may be considered for docking molecular solutes, or a molecule and a surface, is to also compute the short-range electrostatic desolvation and hydrophobic desolvation terms, in addition to the electrostatic interaction term, by additionally setting edfct and hdfct to non-zero values, see below. This combination is particularly relevant for the docking of solutes that have weak electrostatic attraction. While it has been applied to a variety of molecular systems, it has not yet been systematically parameterized for general application to docking to generate diffusional encounter complexes (or to computing bimolecular association rate constants).
- ---For simulating the diffusion of multiple proteins, set epfct=0.5, edfct=0.36 (with electrostatic desolvation grids computed with iostr= 0 in ed.in), hdfct=-0.0013 (see below on modifying this value), and lj_rep_fct=1.0. The same settings should be applied for simulating the diffusion of multiple proteins in the presence of a non-metal surface.
-
epfct [float] (0.0) : factor by which the electrostatic
potential grid values read in are to be multiplied.
Should be 0.5 for all calculations to compute the electrostatic interaction force if no modifications are
needed. (This setting is equivalent to setting 1.0 in previous versions
of SDA)
-
edfct [float] (0.0) : factor by which the electrostatic
desolvation potential grid values are to be multiplied to compute the electrostatic desolvation forces.
The value 1.0 corresponds to the uncorrected
electrostatic desolvation penalty for a charge in a
solvent due to the low dielectric cavity of a solute
treated as a collection of van der Waals spheres. More
appropriate values of this factor can be derived by comparing SDA
calculated energies of recorded complexes with energies
calculated with more accurate methods (by solving the
finite difference Poisson-Boltzmann equation, for
example). The accuracy with which the parameterization reproduces Poisson-Boltzmann energies and forces will vary depending on the solute properties, their proximity and the environmental conditions. One of two parameterizations of the electrostatic desolvation forces should be used:
(1) edfct=1.67 with solute interior
dielectric constant set to 4 and the dielectric
boundary between the solute interior and high
dielectric solvent defined by the van der Waals surface
of the solute. In this case, the electrostatic desolvation term is dependent on ionic strength and is computed with the same ionic strength (iostr) assigned in the grid preparation in ed.in as for the Poisson-Boltzmann electrostatic potential calculations. This parameterization was derived to reproduce Poisson-Boltzmann interaction energies of a set of protein-protein diffusional encounter complexes and used for the computation of protein-protein diffusional association rate constants, see:
Gabdoulline, R. R.; Wade, R. C., J. Mol. Biol. 2001,
306, 1139 - 1155
Article
link.
N.B. For simulating a protein-metal surface system using the ProMetCS force field, set edfct to 2x1.67 =3.34 to ensure that the contribution to the electrostatic desolvation term with the image charge model is accounted for (this is equivalent to using edfct=1.67 between two standard solutes).
(2) edfct=0.36 with solute interior
dielectric constant set to 4 and the dielectric
boundary between the solute interior and high
dielectric solvent defined by the van der Waals surface
of the solute. In this case, the electrostatic desolvation term is not dependent on ionic strength and is computed with the ionic strength zeroed (iostr = 0) in the grid preparation in ed.in. Note that the electrostatic interaction term remains dependent on the ionic strength used in the Poisson-Boltzmann electrostatic potential calculations. This parameterization was derived to provide a simple protocol for computation of protein-protein electron transfer rates at different ionic strengths, see: Gabdoulline R.R., Wade. R.C. JACS, 2009, 131, 9230
Article
link, and is employed in webSDA. In subsequent SDAMM multiple molecule studies, this parameterization has been used with the interior dielectric constant set to 2, see e.g. Mereghetti P.M., Gabdoulline R.R., Wade R.C. Biophys. J., 2010, 99:3782-3791.
Article
link.
-
hdfct [float] (0.0) : factor by which the hydrophobic
desolvation potential grid values are to be multiplied to compute the short-range attractive nonpolar interaction forces.
In the SDA program, hydrophobic desolvation energies
are calculated by multiplying the solvent
accessibilities of the atoms of one of the solutes by
the value of the hydrophobic desolvation potential grid
calculated for the other solute. The hydrophobic desolvation potential grid should be
calculated with parameters assigned so that the sum of contributions from all
accessible atoms gives the buried solvent accessible
area, i.e. with a=3.1 Å, b=4.35 Å, and factor=0.5 in hd.in.
The hdfct factor has units of kcal/mole/Å2 and converts buried area into hydrophobic desolvation energy.
Realistic values for this factor based on analyses of buried surface area and known binding affinities of protein-protein complexes lie in the range
range -0.0050 to -0.060 kcal/mole/Å2
(-5 to -60 cal/mole/Å2).
In the study of plastocyanin-cytochrome F electron transfer, in which the hydrophobic desolvation forces were first introduced into SDA, the best agreement with experiment was obtained when hdfct was assigned a value of -0.019 kcal/mole/Å2. The more negative hdfct is, the more favorable the interaction between the solutes and the longer the residence times of the solute complexes: this can lead to the problem of the simulations becoming very computationally demanding. In this study, values of hdfct from -0.013 to -0.019 kcal/mole/Å2 were used, see:
Gabdoulline R.R., Wade. R.C. JACS, 2009, 131, 9230
Article
link.
In subsequent publications describing simulations with hydrophobic desolvation forces, values in the range
from -0.005 to -0.019 kcal/mole/Å2 have been used, with most studies done with hdfct = -0.013 kcal/mole/Å2. However, note that for simulations of protein-gold surface interactions with the ProMetCS force field (which also includes metal desolvation and Lennard-Jones terms), hdfct = -0.005 to -0.0065 kcal/mole/Å2 is used, see: Kokh et al. JCTC, 2010, 6, 1753
Article
link.
-
ljfct [float] ( 0.0) : factor by which the Lennard-Jones
potential grid values are to be multiplied when using the ProMetCS force field.
-
lj_rep_fct [float] ( 0.0) : factor by which the soft-core
repulsion (repulsive Lennard-Jones) grids are
multiplied to compute the soft-core repulsion forces. It was introduced for use with sdamm for multiple molecule simulations, and replaces the use of
the exclusion grids. If the repulsive Lennard-Jones grids are computed with an assignment of factor=4096 in
ljrep.in, then lj_rep_fct=0.0156 should be used (see also the Preparation tools documentation).
-
oneway_surf_charge [bool] (0) : if set to 1, the electrostatic interactions between a surface and a solute will be computed in one direction with the grid for the surface interacting with the (effective) charges on the solute. If set to 0 (default), the electrostatic interactions are computed in both directions (average of surface grid with solute charges and grid charges with solute grid)
-
epfct_oneway_surf [float] (1.0) : scaling factor for the one-way electrostatics which should be set to 1.0 (default).
-
Other parameters
-
ignore_exclusion [bool] ( 0 ) : if set to 1, the excluded volume check
will not be performed to prevent solute overlaps. In this
case, soft-core repulsion should be used instead.
-
restart [string] ( empty ) : input file from which the initial
solute positions are read (sda_energy, sda_koff,
sdamm). If this is commented, the positions in the
input pdb files will be used as the starting positions
instead (for sda_koff)
-
account_occurency [bool] ( 0 ) : Only in the case of
sda_koff, if set to 1, the trajectory
will restart n_occurency * nrun times
with the initial position given by the complex.
-
novers [integer] ( 150 ) : number of overlaps allowed before
making a boost. Recommended value: 100-200. With a
value in this range, boosts are usually not necessary
unless the interactions are very strong. This can be
the case if a very small probe size
(probep) is chosen. Many boosts
indicate erroneous input.
-
rboost [float]( 1.0 ) : boost distance (in Å) to move the
solutes apart when more than novers
moves were not accepted due to overlaps. Boosting
is introduced to avoid infinite trajectories and can
influence simulation results because trajectories are
modified by boosts. The number of boosts is reported in
the output log.
-
Parameters specific to the ProMetCS force field
-
correction_image_charge
[float]( 0.0 ) : a non-zero positive number means that
the analytical correction will be applied in computing
the image charge electrostatics. This parameter is also
used to re-scale effective charges when image-charge
electrostatics is computed analytically (at small
distances from the surface when the solute-surface
interface is partially desolvated).
If the protein effective charges do not deviate
strongly from the test charges in magnitude (this is
usually the case for proteins), use
correction_image_charge = 1.
However, if the effective charges of a solute are too
small (as for the case of DNA simulation), the
parameter correction_image_charge enables correcting
back (particularly, to increase) solute charges to test
charges when they are used for the calculation of
image-charge electrostatics at small solute-surface
distances
Note, that the parameter image_charge
= 1 must be defined to use this function.
-
If correction_image_charge > 0, the dielectric
constant for the case of atomic cavities in the
solute molecule close to the surface is scaled as
follows:
ε=4.0+A*z*z+exp(-z/B - C) for 2.5 < z < 5.5 Å,
where :
- A ; default value: A = 0.8 1/Ų
- B ; default value: B = 0.39 1/Å
- C ; default value: C = 10.4
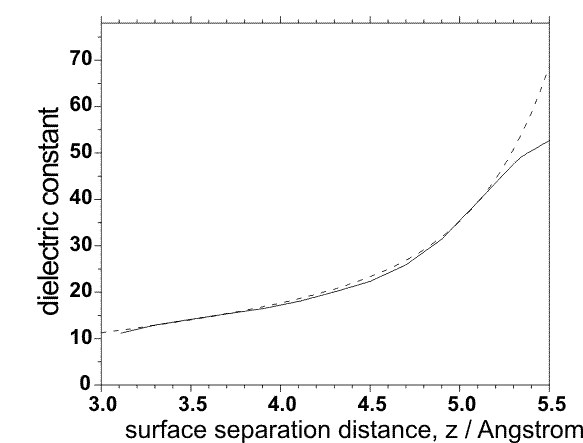
These parameters have been derived from MD
simulations to compute the total electrostatic
interaction energy for a test charge atom as a
function of the distance from a gold surface and are
hard-coded in the file mod_compute_image_charge.f90
(ep_scaleA, ep_scaleB and ep_scaleC)
-
desolv_image_charge
[integer](
0 ) : indicates if
the image charge calculation includes the electrostatic
desolvation term.
If it is equal to 0, it is deactivated but will be
automatically switched on if the electrostatic image
charge is computed.
To force the deactivation of this term in this case,
use desolv_image_charge = -1.
[Back to Main documetation]
Imprint/Privacy