This example shows the implementation of the potential of mean force (PMF) calculation in SDA 7 with a system of a three-histidine tripeptide (3HIS) tripeptide on an Au(111) surface. Details are given in : Ozboyaci, M., Martinez, M. and Wade, R.C. An Efficient Low Storage and Memory Treatment of Gridded Interaction Fields for Simulations of Macromolecular Association.J. Chem. Theory Comput. (2016), 59:7598-7616. 10.1021/acs.jctc.6b00350
The PMF code has been implemented in SDA 7 and tested only for ProMetCS force-field (See aubs example).
This example can be found in directory examples/au3his/.
| prepare_grids_and_ecm/ | Directory for generating grids and effective charges needed for running the simulation |
| data_grid/ | Data files (pdb and input files) needed for running the examples |
| run_hits/ | Results of the example run script for protein-metal surface docking |
| run/ | Directory to run the PMF calculations |
| doc/ | This documentation |
A number of sampling parameters should be given in the input file. Refer to the input page for detailed descriptions of the parameters.
- Number of angles (for each of the Euler angles) to be used for the sampling of the rotation of the rigid body structure (
mentth,mentfi,mentom). - Rectangular area of the surface to be covered in calculations (
mentxandmentyin angstrom units), at the exception of the surface. - The distance from the center of geometry of the protein to the center of the surface in z direction (
pstartandpfinish) and the sampling interval (dz) in angstrom units.
Assure you have compiled all the executables and tools in sda_flex/bin/ first or refer to the compilation section.
First, run the script run-au3his_ed_hd_ecm_lj.sh from the prepare_grids_and_ecm/ directory to generate the grids necessary for this example.
Then go into the run/ directory that contains the input file and run the shell script
For the PMF calculations :
cd run/
./run_sda_pmf.sh
You can check that output files are similar to the ones generated in the *_hits directory.
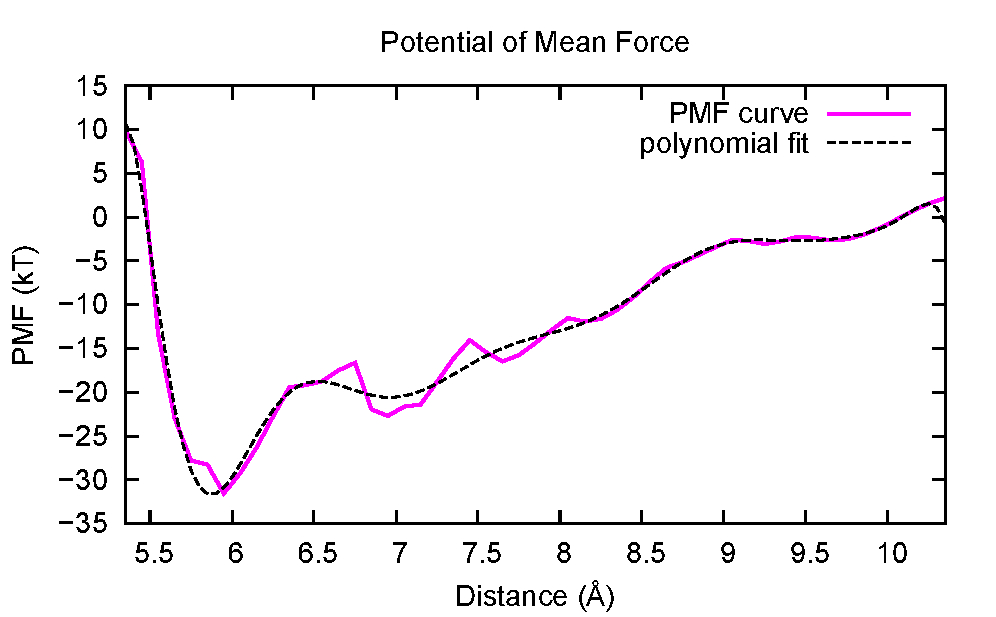
Calculations will produce a file called pmf.dat that contains the PMF values at the distances from the surface based on the dz value.
Using gnuplot plotting program with the input file make_plot.gp a graphics file named pmf_au3his.eps will be obtained. Using a graphics viewer that supports EPS format, the following figure can be obtained.